The expression of bitter taste receptors (T2Rs) in the porcine gastrointestinal tract epithelium and smooth muscle
N. Tang A , V. Ting A , M. Fu A and E. Roura A BA The University of Queensland, Brisbane, QLD 4072.
B Corresponding author. Email: e.roura@uq.edu.au
Animal Production Science 57(12) 2420-2420 https://doi.org/10.1071/ANv57n12Ab108
Published: 20 November 2017
The expression of bitter taste receptors (T2Rs) was originally discovered in taste sensory cells of the tongue’s taste buds. However, in recent years, it has been found that T2Rs are expressed beyond the oral cavity in mammals including pigs where they seem to play a fundamental role orchestrating the hunger-satiety cycle (Roura et al. 2016). However, little is known about their specific expression profile across tissues. This project aimed to investigate the expression of a subset of porcine T2Rs throughout the epithelium of the gastrointestinal tract (GIT), to quantify the abundance and localisation of the genes of interest expressed, in particular in the stomach, duodenum and oesophagus.
Six Large-White male piglets with 10.2 ± 0.53 kg bodyweight were used to perform the intestinal gene expression analysis. Five porcine bitter taste receptors T2R1, T2R4, T2R7, T2R10, T2R20 and T2R39 were selected based on their high gene sequence homology with the human orthologues and their affinity to several compounds (i.e. caffeine, quinine, amarogentin and saccharin) known to be bitter to humans and pigs. Epithelium layers and smooth muscle layers in the stomach, duodenum and oesophagus were separated and analysed for the expression of the bitter taste receptor genes (Tas2rs). Total RNA was extracted from the tissue samples using the TRIZOL-chloroform method. RNA purification, cDNA synthesis and real-time qPCR assays using SYBR green were performed. The relative gene expression levels were estimated using the Pfaffl method (Pfaffl 2001) taking into account the cycle threshold values of both the candidate genes and of the two reference genes ACTB3 and GAPDH. GraphPad Prism 7 software (GraphPad Software Inc., La Jolla, CA, USA) was used to analyse the gene expression data using a one-way ANOVA and Fischer’s least significant difference (l.s.d.) analysis for multiple comparisons.
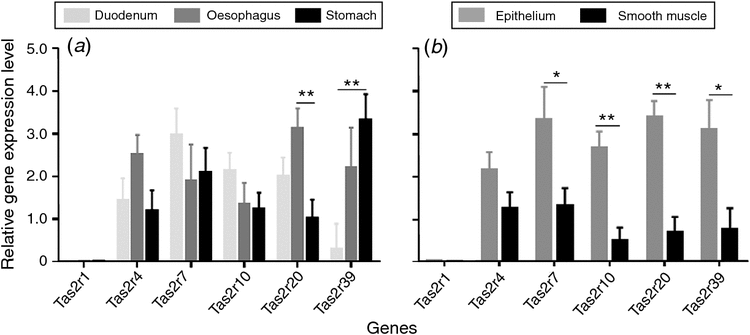
The porcine Tas2r20 gene had a higher expression in the oesophagus than in the duodenum and stomach (P < 0.01; Fig. 1a). Tas2r39 resulted in higher expression rates in the stomach compared to the oesophagus and duodenum (P < 0.01; Fig. 1a). Porcine Tas2r7 and Tas2r39 genes both had a greater relative expression in the epithelium than the smooth muscle layer (P < 0.01; Fig. 1b). Porcine Tas2r10 and Tas2r20 genes both had a greater relative expression in the epithelium than the smooth muscle layer (P < 0.05; Fig. 1b).
The results confirmed the expression of porcine Tas2r genes beyond the oral cavity into the gut with the level of expression being gene and tissue specific. In addition, our data shows that the expression of the genes of interest occurs preferentially in the epithelial cell layer of the GIT. These results are relevant in the context of understanding the functionality, including tissue specificity, of food-borne compounds relevant to appetite enhancing or inhibition.
References
Pfaffl M (2001) Nucleic Acids Research 29, e45| Crossref | GoogleScholarGoogle Scholar |
Roura E, Koopmans S, Lallès J, Le Huerou-Luron I, de Jager N, Schuurman T, Val-Laillet D (2016) Nutrition Research Reviews 29, 60–90.
| Crossref | GoogleScholarGoogle Scholar |