A novel polymorphism in the 5′ UTR of HvDEP1 is associated with grain length and 1000-grain weight in barley (Hordeum vulgare)
Calum Watt A B E , Gaofeng Zhou A B , Tefera Tolera Angessa A B , David Moody C and Chengdao Li
A B E , Gaofeng Zhou A B , Tefera Tolera Angessa A B , David Moody C and Chengdao Li  A B D E
A B D E
A Western Crop Genetics Alliance, Murdoch University, 90 South Street, WA 6150, Australia.
B Western Australian State Agricultural Biotechnology Centre, Murdoch University, 90 South Street,WA 6150, Australia.
C Intergrain Pty Ltd, 19 Ambitious Link, Bibra Lake, WA 6163, Australia.
D Department of Primary Industries and Regional Development, WA, Australia.
E Corresponding authors. Email: C.li@murdoch.edu.au; calumwatt12@outlook.com
Crop and Pasture Science 71(8) 752-759 https://doi.org/10.1071/CP20169
Submitted: 22 May 2020 Accepted: 5 August 2020 Published: 18 August 2020
Journal Compilation © CSIRO 2020 Open Access CC BY-NC-ND
Abstract
The gene HvDEP1, on barley (Hordeum vulgare L.) chromosome 5H, encodes a γ-subunit of the heterotrimeric G-protein complex and was previously determined to be a candidate gene underlying a major quantitative trait locus for grain length. In the present study, we identified a 9 bp indel (insertion–deletion mutation) at position –84 bp from the start codon within a reported upstream open reading frame located in the 5′ UTR (untranslated region) and developed a diagnostic molecular marker. We also identified a 13 bp indel (–514 bp) in linkage disequilibrium that bridges an important regulatory motif. Using a doubled-haploid population and a barley diversity panel, we were able to show that the effects of these indels were environmentally stable and consistently delineated phenotypic groups based on grain length and 1000-grain weight. Genotypes represented by deletions at these two positions relative to the reference cv. Morex had consistently shorter grains, by 3.69–3.96%, and lower 1000-grain weight, by 2.38–4.21%, in a doubled-haploid population studied. Additionally, a diversity panel was tested but consistent differences were observed only for grain length, reinforcing literature indicating the importance of this gene for grain-length regulation. The frequency of the longer and heavier grained reference allele was higher in modern cultivars, suggesting that indirect selection for longer grain may have occurred through direct selection for grain yield via grain-weight improvement. These results indicate that grain length and 1000-grain weight in barley can be manipulated by targeting variation in gene promoters through marker-assisted selection.
Additional keywords: cis-element, DEP1, grain length, promoter, TGW, uORF.
Introduction
As the fourth most important cereal crop grown globally, improving the yield and quality of barley (Hordeum vulgare L.) represents an important focus of breeding programs. Grain yield in barley is a product of number of grains per area and individual grain weight. In rice (Oryza sativa L.), individual grain weight is influenced by grain length, changing the physical appearance of the grain, which is part of the culinary quality (Li et al. 2019). As such, many loci and genes have been identified in rice as contributing to variations in grain length; for example, qGL3 is associated with increased 1000-grain weight (TGW) owing to a significant increase in grain length with little to no interaction with environment (Wan et al. 2006; Zhang et al. 2012). A GNAT-like protein encoded by OsGW2 increases grain length and weight through spikelet hull enlargement and accelerated rate of grainfill (Song et al. 2015). Rice qPE9-1/DEP1 has been manipulated to varying extents, with multidimensional effects on grain length and weight dependent on the specific genetic background and allele (Li et al. 2019). This multidimensional effect of DEP1 has also been identified in barley (Wendt et al. 2016). Wan et al. (2006) concluded that smaller effect quantitative trait loci (QTLs) influencing grain length were less environmentally stable than qGL3 because of environmentally induced epistasis between these loci, unlike the major effect QTL.
Breeding for high levels of grain plumpness, as measured by retention of grain on a slotted screen and often referred to as screenings, is a key focus in barley breeding, but grain plumpness has not generally been selected for by deliberately manipulating grain length. Understanding the relationship between grain plumpness, grain length and grain weight when breeding for yield in barley is critically important to breeding programs attempting to select for improved grain yield while simultaneously at least maintaining industry-acceptable levels of grain plumpness. Grains of landrace barleys and barley progenitor species (Hordeum vulgare ssp. spontaneum) are typically much longer and less plump than those of modern cultivars. However, there is sufficient variation in modern cultivars to warrant population development for the purposes of improving our understanding of the genetic basis of grain size in barley and increasing the rate of introgression of positive grain-size alleles into elite backgrounds, providing the negative relationship between grain length and plumpness can be broken (Schnaithmann and Pillen 2013).
In a previous study, a major locus on barley chromosome 5H was found to have significant influence on grain length. This particular locus was able to explain 21.6% of the phenotypic variation for this grain length, and was identified in a doubled-haploid (DH) population derived from two elite Australian barley cultivars, Vlamingh and Buloke. A candidate gene identified underlying this locus, HvDEP1, represents a promising target for further genetic and molecular characterisation (Watt et al. 2019). HvDEP1 shares 94.8%, 81.2% and 88.8% sequence identity with wheat (Triticum aestivum L.), maize (Zea mays L.) and rice orthologues, respectively, making it a suitable target for further molecular characterisation.
In barley, our understanding of which genes contribute to variation in grain size is limited and is based primarily on the collinearity of gene sequence and function between the major cereal crop species. In this study, we report the identification of novel polymorphisms in the promoter region of HvDEP1 that are diagnostic for variation in grain length and TGW. HvDEP1 encodes a plasma-membrane-associated Gγ subunit of the G-protein signalling pathway that dimerises with a Gβ subunit and interacts with the Gα subunit to perform a variety of signalling functions (Wendt et al. 2016; Sun et al. 2018). Several polymorphisms identified in the promoter region of HvDEP1 in the present study are believed to drive variation in gene expression, resulting in changes in grain length and TGW in both a DH population and a natural population included in this study. We further investigate the prevalence of promoter polymorphism in modern barley cultivars and its relevance for barley breeding.
Materials and methods
Plant material and phenotypic analysis
Two different sets of barley germplasm were used in this study: (i) a DH population consisting 246 lines in total, derived from two Australian two-rowed barley varieties, cv. Vlamingh and cv. Buloke; and (ii) a diversity panel comprising 64 barley cultivars, landraces and breeding lines (see Supplementary Materials table S1, available at the journal’s website). Field trials were partially replicated and arranged in complete blocks. Current commercial varieties acted as controls. The DH population was grown during 2007 under irrigation and rainfed conditions at Horsham, Victoria, and during 2017 under irrigated conditions at South Perth, Western Australia. The number of lines represented ranged from 228 to the full population across the three trials. The barley germplasm diversity panel was grown during 2018 at South Perth in trials across five times of sowing (TOS) from mid-April to mid-August, with monthly intervals between consecutive sowings. These five TOS represent different growth and maturity environments driven largely by variation in temperature and photoperiod (supplemental irrigation was supplied when necessary to avoid drought). These TOS trials were used to interrogate the relationship between grain length and TGW across varying environments in the diversity panel. Grain length and TGW were measured with an SC6000R digital image analyser (Next Instruments, Condell Park, NSW). Measurements were performed on a subsample of 300–400 grains from each harvested plot.
Molecular marker development and sequencing
Genomic DNA for both sets of barley germplasm was extracted from young leaf tissue. Additionally, DNA was extracted from 1111 modern cultivars and 96 wild barleys for analysis of allele frequency and interrogation of linkage disequilibrium between identified polymorphisms; of theses 1207 lines, of the wild barleys, only 64 had phenotypic data available. Extraction of DNA followed an amended method of Ahmed et al. (2009) described by Watt et al. (2019), the main difference represented by a protein degradation rather than precipitation. Previous research associating HvDEP1 with grain length indicated that the parents of the DH population should be sequenced to identify any potentially diagnostic polymorphisms that might be driving variation in grain length (Wendt et al. 2016; Watt et al. 2019). The coding and promoter region (1.6 kb upstream) of HvDEP1 was downloaded from the BarleyVar database, an in-house database of the Western Barley Genetics Alliance, Murdoch University, developed based on the chromosome conformation capture ordered sequence of the reference variety cv. Morex (Mascher et al. 2017). Primers were then designed spanning the entire main open reading frame of HvDEP1, including 2 kb upstream and downstream start and stop codons, respectively, using Geneious Prime 2019.2.1 software (https://www.geneious.com/prime/) (table S2). Primers pairs were designed ensuring that amplicons overlapped.
High-quality genomic DNA was isolated from the DH population, their parent cultivars and the diversity panel as described previously (Watt et al. 2019). PCR reactions were performed in a total volume of 10 μL containing 1 µL 10× buffer and GC buffer, 0.25 mM dNTPs, 0.2 µM each primer, 50 mM MgCl2, 50 ng genomic DNA, and 0.2 U Taq-polymerase. The thermocycler protocol was as follows: 95°C for 3 min, 38 cycles of 94°C for 20 s, 55–57°C (primer-dependent) for 20 s, 72°C for 20 s, and a final extension at 72°C for 5 min. Amplicons for sequencing were separated in 1% agarose gels in 1× TAE buffer, and correct bands were excised from the gel under UV light. Amplicon DNA was purified from gel fragments by using a Wizard SV Gel and PCR Clean-Up System (Promega, Madison, WI, USA) and adjusted to 100 ng μL–1 for sequencing.
One-eighth Sanger sequencing reaction concentrations were made for each purified amplicon in 10-µL volumes containing 1 µL dye terminator mixture, 1.5 µL 5× sequencing buffer, 3.2 pmol primer (forward or reverse) and 50 ng DNA amplicon, and made up to final volume with sterile water. The thermocycler protocol was as follows: 96°C for 2 min followed by 25 cycles of 96°C for 10 s, 56°C for 5 s, and 60°C for 4 min. Post-reaction purification was then performed by using an ethanol– EDTA–sodium acetate precipitation step followed by sequencing in an Applied Biosystems 3730 Genetic Analyzer, 96-capillary array (Thermo Fisher Scientific, Waltham, MA, USA). Overlapping sequences were aligned using Geneious Prime 2019.2.1 software. Primer pairs based on identified polymorphisms between the two parents were developed and tested on the DH population and the diversity panel. Amplicons were separated in 2% agarose gels in 0.5× TBE buffer and visualised under UV light.
In silico analysis of HvDEP1 promoter
A 1.6-kb region upstream of the HvDEP1 start codon was amplified from both cv. Vlamingh and cv. Buloke (table S2). Both sequences were searched for cis-regulatory elements in the plant promoter database PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) and compared with the reference sequence of cv. Morex (Lescot et al. 2002). In silico protein modelling prediction was performed using I-TASSER software (https://zhanglab.ccmb.med.umich.edu/I-TASSER/) by inputting the upstream open reading frame (uORF) sequence.
RNA isolation, cDNA synthesis and RT-qPCR
Total RNA was isolated from 1-cm developing inflorescences of cvv. Vlamingh and Buloke by using TRIsure following the manufacturer’s protocol for plant tissues (Bioline, London). Genetic and functional sequence assembly of the barley reference cultivar (Morex) has defined the expression profile of HvDEP1, with the highest relative expression exhibited in the developing inflorescences at 1 cm (Mayer et al. 2012; Colmsee et al. 2015). Four biological replicates were collected from individual plants grown under glasshouse conditions. RNA was precipitated with isopropanol and resuspended in water treated with DEPC (diethyl pyrocarbonate). Quality and quantity of RNA was determined on a NanoDrop One/One spectrophotometer (Thermo Fisher Scientific). First-strand complimentary DNA (cDNA) was synthesised from 1 µg RNA using the SensiFASTcDNA Synthesis Kit (Bioline) following the manufacturer’s protocol. Reverse transcription quantitative real-time PCR (RT-qPCR) reactions were performed by using the SYBR Green detection chemistry in 10-µL reaction volumes containing 5 µL SYBR Green Supermix (Bio-Rad, Hercules, CA, USA), 10× forward and reverse primers (table S2) and 100 ng cDNA template, and made up to final volume with sterile water. Each biological replicate had three technical replicates with the reaction performed in a ViiA-7 Real-Time PCR System, and data were analysed using QuantStudio software (Thermo Fisher Scientific). Relative expression was calculated using the 2−ΔΔCT method, and HvActin was used for expression normalisation.
Results
Analysis of HvDEP1 promoter region
The HvDEP1 gene, including 1.6 kb upstream and 1 kb downstream was sequenced from both DH population parent cultivars (Vlamingh and Buloke). No polymorphisms were identified within any of the five exons comprising this gene or the 3′ UTR (untranslated region) sequenced.
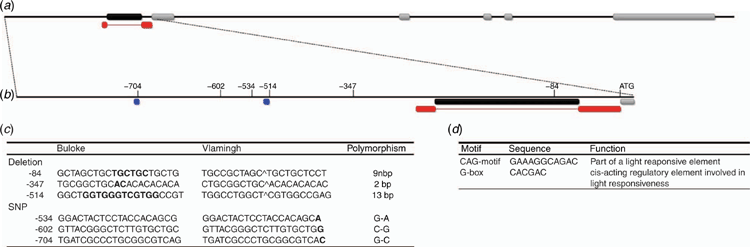
Analysis of the promoter region identified several polymorphisms, of note two large indels (insertion–deletion mutations) at positions –84 bp and –514 bp from the start codon. The indel at position –84 bp was a 9 bp polymorphism present in a reported uORF encoding for a transcript of 70 amino acids located within the long 297 bp 5′ UTR. Additionally, the 13 bp indel was located –514 bp upstream of the transcription start site at a position represented by a G-box regulatory motif (Fig. 1). In both instances, cv. Vlamingh represented the mutant allele with a deletion in a repetitive sequence of 5′-GCTGCTGCT-3′ in the uORF and a deletion of 5′-GGTGGGTCGTGGC-3′ at the position of a G-box. Interestingly, screening a large collection of modern cultivars and wild barley accessions determined that the cv. Buloke allele was over-represented at 82% in the modern cultivars and 66% in the wild barleys. An additional single-nucleotide polymorphism (SNP) (G–C) was located at position –704 bp, which in cv. Vlamingh was responsible for the loss of a CAG-motif (Fig. 1). Other polymorphisms were identified; however, they did not coincide with cis-regulatory elements and were not further explored (Fig. 1b).
In silico sequence transcription indicated a truncated protein of 67 amino acids encoded by the cv. Vlamingh uORF compared with cv. Buloke and previous reports (Bélanger et al. 2014). Furthermore, in silico protein structure prediction using I-TASSER software indicated the loss of a stabilising β-strand as a result of this 9 bp deletion that truncated the cv. Vlamingh predicted peptide (Yang et al. 2015).
Expression profile of HvDEP1
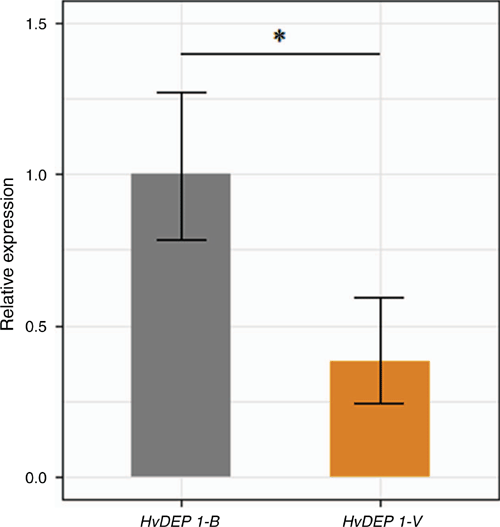
The relative expression profile of HvDEP1 was compared between the DH population parents in the developing inflorescences (1 cm) by using RT-qPCR and gene-specific primers DEP1-cDNA-RT normalised against HvActin (table S2). There was a significant 2.6-fold reduction in the relative expression of HvDEP1 in cv. Vlamingh compared with cv. Buloke (Fig. 2). Surprisingly, uORFs are reported to reduce downstream gene expression primarily through post-transcriptional control, although this finding does not corroborate the reduced grain length and gene expression pattern of cv. Vlamingh, which represents the mutant allele in this instance.
|
Phenotypic validation of promoter polymorphism
Molecular markers were designed for the two indels and a SNP located within the regulatory elements of the promoter region; however, no recombinant individuals were identified by using these markers in the DH population. Therefore, we chose to genotype the diversity panel with only the DEP1-uORF marker. Two allelic groups were demarcated based on the polymorphism identified in the 5′ UTR in both sets of germplasm. These two groups, designated as ‘HvDEP1-V’ and ‘HvDEP1-B’, representing cv. Vlamingh and cv. Buloke allelic groups, respectively, exhibited consistent differences in grain length and TGW between them across multiple contrasting environments. In the DH population, grain length and TGW were highly significantly correlated in each environment (r = 0.31–0.71; P < 0.001), with significant correlations observed between environments (Fig. 3a). Additionally, the two allelic groups exhibited consistently significant differences for TGW and grain length. Grain length was 3.69–3.96% greater for the HvDEP1-B allelic group than HvDEP1-V with a range of 8.39–8.76 mm (Table 1). Consistent with the significant positive Pearson’s correlation between grain length and TGW, HvDEP1-B had significantly higher TGW than the alternative allele, by 2.38–4.21%.
As expected for a diversity panel, there were large phenotypic variations within each allelic group delineated by their DEP1-uORF genotype driven by the strong genetic diversity present within this panel (table S1). For example, within the HvDEP1-B allelic group, grain length ranged from 7.79 to 10.27 mm for the April TOS trial. Average grain lengths for the HvDEP1-B allelic group were significantly greater across April, June and August TOS trials, ranging from 8.36 to 8.75 mm, representing a 3.79–5.08% increase on the HvDEP1-V allelic group (Table 1). There was a significant difference in TGW for the April TOS only, with the HvDEP1-B group 8.23% heavier than HvDEP1-V. Grain length and TGW were significantly positively correlated with each other in each TOS trial apart from August (Fig. 3b). There was no clear trend in the diversity panel with later TOS and trait variation; in fact, the May TOS trial had the lowest TGW and shortest grains. The relatively late July TOS resulted in longer grains but lower TGW than the TOS that would be considered optimal (i.e. April): 8.87 ± 0.07 mm and 50.3 ± 0.86 g compared with 8.54 ± 0.03 mm and 58.87 ± 0.39 g. Principal component analysis of the diversity panel indicated that neither grain length nor TGW sufficiently discriminated on the basis of TOS, suggesting that either a reduced influence of environment on these two traits or the high degree of background genetic variability led to varying genotype performance with later TOS (Supplementary fig. S1B). Indeed, some varieties tended towards longer grains with later TOS, whereas some tended towards shorter grains, although this did not necessarily reflect origin of the germplasm (table S1). Principal component analysis indicated that TGW was more discriminating than grain length in the DH population; however, grain length did vary significantly between the irrigated (8.6 mm) and rainfed (8.23 mm) Horsham trials, and between the 2017 (8.53 mm) and rainfed Horsham trials, suggesting that water limitation may be the leading driver of the grain-length variation observed.
Discussion
Identification of candidate genes that contribute to variation in grain size and weight is in its infancy in barley, and more so is the identification of novel polymorphisms that are diagnostic for trait variation across multiple environments and therefore appropriate for marker-assisted selection (MAS). Previous studies have identified polymorphisms in DEP1 as being responsible for variation in grain length, weight and plant architecture in rice (Huang et al. 2009; Sun et al. 2018; Li et al. 2019). Furthermore, previous studies in barley have mapped major grain-length loci, with one study fine-mapping a major locus on chromosome 5H and identifying HvDEP1 as a promising candidate gene warranting further research (Wendt et al. 2016; Watt et al. 2019). In the present study, we were able to associate a novel HvDEP1 promoter polymorphism to variation in grain length and TGW in two distinct sources of barley germplasm across multiple environments, indicating the stability of the allelic effect. In the present research, no polymorphisms were detected in the main coding region of HvDEP1 or the 3′ UTR. Previous research has not addressed the role of the promoter region in expression of HvDEP1; therefore, we addressed this deficiency by sequencing a 1.6-kb promoter region immediately upstream of the start ATG codon and identified six polymorphisms between cv. Vlamingh and cv. Buloke. Of these identified variants, three polymorphisms required further interrogation because they were located in important regulatory elements. A 9 bp deletion was identified in a reported uORF at position –84 bp, a regulatory element conserved between the major cereal-crop species and reported to be involved in post-transcriptional regulation (Bélanger et al. 2014). An additional deletion of 13 bp was identified in cv. Vlamingh bridging a G-box regulatory element located at –514 bp. Last, a G–C SNP at –704 bp in cv. Vlamingh resulted in the deletion of a CAG-motif. Molecular markers were designed for all polymorphisms, but owing to linkage disequilibrium of all identified polymorphisms in the promoter region, only the 9 bp indel marker was used for genotyping purposes presented here.
Allelic groups delineated by the 9 bp indel marker consistently exhibited significantly different grain length and TGW in numerous environments. A DH population derived from cvv. Vlamingh and Buloke, with contrasting grain lengths, was phenotyped in three independent environments. DH lines with the HvDEP1-B allele had significantly longer grains and higher TGW in all environments tested (Table 1). Despite the promise of this marker for delineating superior allelic groups, testing within a DH population with limited genetic variability is not indicative of overall marker performance in germplasm with diverse genetic backgrounds. As such, a diversity panel comprising commercial cultivars, barley landraces and breeding lines was screened with this marker. Despite the diverse origins of this material, the marker was able to delineate two allelic groups with contrasting phenotypes. Those matching HvDEP1-B had significantly longer grains in three environments tested than those matching HvDEP1-V, whereas TGW was significantly different for the April TOS trial only, which is likely to be driven by the extended duration of grainfill compared with later TOS (Table 1). Despite the lack of significant differences, the correlation between grain length and TGW was significantly positive in four of five TOS trials (Fig. 3b), supporting the notion that a longer grain contributes to a heavier grain as observed in rice and wheat. However, a greater number of genotypes with phenotypic data and further contrasting environments would have improved our interpretation of marker effects, trait correlation and environmental stability (Wang et al. 2018; Ying et al. 2018). The consistent relationship between the promotor polymorphism and grain length across environments, showing a stable influence of promoter polymorphism, indicates that grain length is much less influenced by environment than is TGW. This is unsurprising given grain length has high heritability compared with TGW, which is controlled by several interrelated factors not exclusive of grain size (Avni et al. 2018; Roy and Shil 2020; Watt et al. 2020). However, the positive correlations between grain length and TGW suggest that this polymorphism would still be useful for MAS improvement of TGW, and hence grain yield, which typically has much lower heritability. Interestingly, Australian National Variety Trial (NVT) data indicate that cv. Buloke has a consistently higher TGW than cv. Vlamingh across contrasting environments, indicating the usefulness of targeting this particular locus through MAS. It should be noted that screenings, an important quality parameter, is highly correlated with grain plumpness. In hotter and drier environments, a plumper variety (typically shorter, i.e. cv. Vlamingh) tends to have lower screenings than a longer grained variety such as cv. Buloke, although NVT data indicate that this contrast becomes less pronounced in favourable environments where source–sink relationships are less limited. In order to improve the potential usefulness of the identified polymorphisms for MAS, additional trials in environments drier and hotter than those tested in the present study would be beneficial, coupled with MAS targeting grain-plumpness loci to address grain-quality variation associated with TGW and screenings.
Expression analysis of HvDEP1 in the developing inflorescences clearly indicated that gene expression was significantly reduced in cv. Vlamingh, by 2.6-fold, compared with cv. Buloke. Extensive research in rice has found that differential expression of qPE9-1/DEP1 induced by gene knockout and overexpression significantly influenced grain length, TGW and plant architecture (Huang et al. 2009; Sun et al. 2018; Li et al. 2019). We were able to identify multiple promoter polymorphisms in linkage disequilibrium in both a DH population and a diversity panel. A molecular marker targeting a 9 bp uORF polymorphism consistently indicated that genotypes harbouring the cv. Vlamingh allele, synonymous with reduced HvDEP1 expression in this instance, exhibited significantly reduced grain length and TGW compared with the reference allele in multiple environments (Table 1). This finding is supported by Li et al. (2019), who found that knockout mutants of OsDEP1 had significantly reduced grain length and TGW compared with wild types. Furthermore, cv. Vlamingh and cv. Buloke have distinct plant architectures, Vlamingh being visibly more erect than Buloke, which has implications for light-harvesting capability, in-crop ventilation and potential tolerance to crop lodging. Unfortunately, the favourable allele for TGW from cv. Buloke is associated with the less erect canopy structure. Similarly, in rice, the allele for reduced grain length at the qPE9-1/DEP1 locus was associated with a more erect plant structure and with negative yield effects (Li et al. 2019).
The absence of two cis-regulatory elements and truncation of the uORF in cv. Vlamingh offer multiple possible explanations as to the molecular processes that have occurred to reduced gene expression significantly and drive phenotypic variation. The first explanation pertains to the presence of a predicted uORF, a cis-regulatory element proven to control gene expression predominantly through post-transcriptional regulation. Upstream ORFs encode for small peptides that are highly conserved and over-represented in the 5′ UTR of regulatory genes such as those that control plant growth, development and response to environment, and the presence of such regulatory elements commonly downregulates translation of the main CDS region (Morris and Geballe 2000; Jorgensen and Dorantes-Acosta 2012). It is therefore unsurprising that we find such a cis-regulatory element in the 5′ UTR of HvDEP1, owing to its role in G-protein signalling (Wendt et al. 2016; Sun et al. 2018). Research assessing the role of uORF-mediated gene expression in plants has focused on the post-transcriptional control through processes such as ribosomal stalling and mRNA decay (Laing et al. 2015; Srivastava et al. 2018). Protein synthesis experiments were not conducted in this study; however, RT-qPCR analysis of HvDEP1 concluded that expression of HvDEP1 in cv. Vlamingh was significantly reduced compared with cv. Buloke. Amino acid sequences of translated uORFs located in the 5′ UTR of HvDEP1 orthologues are highly conserved, indicating that the translated peptide is the active element involved in gene expression control and not the CDS transcript itself (Bélanger et al. 2014). However, mRNAs undergo several different quality-control processes to protect a cell from translation of aberrant RNAs and potentially toxic or dysfunctional peptides (Karamyshev and Karamysheva 2018). Another possible explanation for reduced gene expression observed in cv. Vlamingh could therefore be related to structural differences in the mRNA or the nascent peptide during translation because of a 9 bp deletion that triggers mRNA decay. In silico analysis of the predicted peptides encoded by cv. Vlamingh and cv. Buloke sequences indicates the loss of a key β-strand secondary structure in the peptide encoded by the cv. Vlamingh uORF. The β-strands are important secondary structures contributing to the stability of a peptide, and research indicates that aberrant proteins can be degraded through non-stop or no-go mediated decay, where mRNAs that lack stop codons are degraded (as in the case for the HvDEP1 uORF) or mRNAs stalled in ribosomal complexes are degraded (Karamyshev and Karamysheva 2018). In the present research, mRNA no-go-mediated decay is unlikely to result in the observed gene expression difference, because GC-rich sequences as characteristics of this 9 bp deletion region are reported targets for ribosomal stalling and no-go decay (Shoemaker and Green 2012). Therefore, if no-go decay were responsible for the observed expression variation, one would expect cv. Buloke to exhibit a significantly reduced gene expression pattern compared with cv. Vlamingh, which lacks this GC-rich segment; this expectation is contrary to RT-qPCR results. Alternatively, non-stop-mediated mRNA decay represents a promising explanation as to how this uORF polymorphism drives gene-expression variation. The truncated peptide translated lacks a stabilising β-strand secondary structure compared to the full-length peptide that may play an important process in protecting the mRNA from non-stop decay.
Aside from potential degradation of the mRNA as a result of an indel located within a reported uORF, two alternative polymorphisms located within cis-regulatory elements upstream of the uORF could be responsible for gene expression and trait variation. A 13 bp deletion and G–C SNP in cv. Vlamingh result in deletions of a G-box and CAG-motif, respectively, upstream of the core conserved TATA- and CAAT-box promoters. Both of the deleted cis-regulatory elements are involved in light-responsiveness pathways (Fig. 1d). CAG-motifs are not well researched; however, Ku et al. (2011) concluded from their study that a mutation of the CAG-motif binding site could change gene expression and was associated with plant architecture in maize. By comparison, G-box regulatory elements have been extensively studied and demonstrated to be essential functional components for downstream gene expression, deletion or mutations of which have been shown to reduce dramatically overall promoter activity and response to stimuli (Menkens et al. 1995; Ravel et al. 2014; Liu et al. 2016; Ramegowda et al. 2017). Variation in grain length and TGW in rice has been shown to be driven by cell proliferation in the developing endosperm and controlled by auxin and cytokinin mediation of DEP1; additionally, G-box regulatory elements have been demonstrated to be essential elements conferring endosperm-specific expression (Ravel et al. 2014; Zhang et al. 2019). Based on the literature, G-box-driven variation in gene expression is preferential to that of the CAG-motif in this particular instance, although progressive 5′ promoter-deletion construct analysis would indicate the influence of both elements on gene expression and trait variation. Therefore, we cannot exclude the CAG-motif deletions or the uORF polymorphism as potential drivers of trait variation based on the current data.
The deletions of two key cis-regulatory elements in the promoter region of cv. Vlamingh offer alternatives to the decay of mRNA as hypotheses regarding the driver(s) of differential gene expression and grain length and TGW variation. The G-protein complexes are positive regulators of grain length and TGW in rice, and reduced expression of HvDEP1, which encodes a γ-subunit of this heterotrimeric complex, is a likely contributor to reduced grain length and TGW of varieties that harbour HvDEP1-V alleles as observed in this study (Sun et al. 2018). A marker designed to target the uORF deletion and in linkage disequilibrium with the other identified polymorphisms was diagnostic for grain length and TGW variation. Furthermore, it appears to be environmentally stable, and the longer and heavier grained allele representative of cv. Buloke (and cv. Morex) over-represented in modern cultivars and wild barleys at frequencies of 82% and 66%, respectively, suggests some selection pressure at this locus.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgement
The authors thank the Australian Grains Research and Development Corporation (GRDC) for additional funding (UMU00049 and UMU1903-003RSX).
References
Ahmed I, Islam M, Arshad W, Mannan A, Ahmad W, Mirza B (2009) High-quality plant DNA extraction for PCR: an easy approach. Journal of Applied Genetics 50, 105–107.| High-quality plant DNA extraction for PCR: an easy approach.Crossref | GoogleScholarGoogle Scholar | 19433907PubMed |
Avni R, Oren L, Shabtay G, Assili S, Pozniak C, Hale I, Ben-David R, Peleg Z, Distelfeld A (2018) Genome based meta-QTL analysis of grain weight in tetraploid wheat identifies rare alleles of GRF4 associated with larger grains. Genes 9, 636
| Genome based meta-QTL analysis of grain weight in tetraploid wheat identifies rare alleles of GRF4 associated with larger grains.Crossref | GoogleScholarGoogle Scholar |
Bélanger S, Gauthier M, Jean M, Sato K, Belzile F (2014) Genomic characterization of the Hordeum vulgare DEP1 (HvDEP1) gene and its diversity in a collection of barley accessions. Euphytica 198, 29–41.
| Genomic characterization of the Hordeum vulgare DEP1 (HvDEP1) gene and its diversity in a collection of barley accessions.Crossref | GoogleScholarGoogle Scholar |
Colmsee C, Beier S, Himmelbach A, Schmutzer T, Stein N, Scholz U, Mascher M (2015) BARLEX: the barley draft genome explorer. Molecular Plant 8, 964–966.
| BARLEX: the barley draft genome explorer.Crossref | GoogleScholarGoogle Scholar | 25804976PubMed |
Huang X, Qian Q, Liu Z, Sun H, He S, Luo D, Xia G, Chu C, Li J, Fu X (2009) Natural variation at the DEP1 locus enhances grain yield in rice. Nature Genetics 41, 494–497.
| Natural variation at the DEP1 locus enhances grain yield in rice.Crossref | GoogleScholarGoogle Scholar | 19305410PubMed |
Jorgensen R, Dorantes-Acosta A (2012) Conserved peptide upstream open reading frames are associated with regulatory genes in angiosperms. Frontiers in Plant Science 3, 191
| Conserved peptide upstream open reading frames are associated with regulatory genes in angiosperms.Crossref | GoogleScholarGoogle Scholar | 22936940PubMed |
Karamyshev AL, Karamysheva ZN (2018) Lost in translation: ribosome-associated mRNA and protein quality controls. Frontiers in Genetics 9, 431
| Lost in translation: ribosome-associated mRNA and protein quality controls.Crossref | GoogleScholarGoogle Scholar | 30337940PubMed |
Ku L, Wei X, Zhang S, Zhang J, Guo S, Chen Y (2011) Cloning and characterization of a putative TAC1 ortholog associated with leaf angle in maize (Zea mays L.). PLoS One 6, e20621
| Cloning and characterization of a putative TAC1 ortholog associated with leaf angle in maize (Zea mays L.).Crossref | GoogleScholarGoogle Scholar | 21687735PubMed |
Laing WA, Martínez-Sánchez M, Wright MA, Bulley SM, Brewster D, Dare AP, Rassam M, Wang D, Storey R, Macknight RC, Hellens RP (2015) An upstream open reading frame is essential for feedback regulation of ascorbate biosynthesis in Arabidopsis. The Plant Cell 27, 772–786.
| An upstream open reading frame is essential for feedback regulation of ascorbate biosynthesis in Arabidopsis.Crossref | GoogleScholarGoogle Scholar | 25724639PubMed |
Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Research 30, 325–327.
| PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences.Crossref | GoogleScholarGoogle Scholar | 11752327PubMed |
Li X, Tao Q, Miao J, Yang Z, Gu M, Liang G, Zhou Y (2019) Evaluation of differential qPE9-1/DEP1 protein domains in rice grain length and weight variation. Rice 12, 5–15.
| Evaluation of differential qPE9-1/DEP1 protein domains in rice grain length and weight variation.Crossref | GoogleScholarGoogle Scholar | 30706248PubMed |
Liu L, Xu W, Hu X, Liu H, Lin Y (2016) W-box and G-box elements play important roles in early senescence of rice flag leaf. Scientific Reports 6, 20881
| W-box and G-box elements play important roles in early senescence of rice flag leaf.Crossref | GoogleScholarGoogle Scholar | 26864250PubMed |
Mascher M, Gundlach H, Himmelbach A, Beier S, Twardziok SO, Wicker T, Radchuk V, Dockter C, Hedley PE, Russell J, et al. (2017) A chromosome conformation capture ordered sequence of the barley genome. Nature 544, 427–433.
| A chromosome conformation capture ordered sequence of the barley genome.Crossref | GoogleScholarGoogle Scholar | 28447635PubMed |
Mayer KFX, Waugh R, Langridge P, Close TJ, Wise RP, Graner A, Matsumoto T, Sato K, Schulman A, Muehlbauer GJ, et al. (2012) A physical, genetic and functional sequence assembly of the barley genome. Nature 491, 711–716.
| A physical, genetic and functional sequence assembly of the barley genome.Crossref | GoogleScholarGoogle Scholar |
Menkens AE, Schindler U, Cashmore AR (1995) The G-box: a ubiquitous regulatory DNA element in plants bound by the GBF family of bZIP proteins. Trends in Biochemical Sciences 20, 506–510.
| The G-box: a ubiquitous regulatory DNA element in plants bound by the GBF family of bZIP proteins.Crossref | GoogleScholarGoogle Scholar | 8571452PubMed |
Morris DR, Geballe AP (2000) Upstream open reading frames as regulators of mRNA translation. Molecular and Cellular Biology 20, 8635–8642.
| Upstream open reading frames as regulators of mRNA translation.Crossref | GoogleScholarGoogle Scholar | 11073965PubMed |
Ramegowda V, Gill US, Sivalingam PN, Gupta A, Gupta C, Govind G, Nataraja KN, Pereira A, Udayakumar M, Mysore KS, Senthil-Kumar M (2017) GBF3 transcription factor imparts drought tolerance in Arabidopsis thaliana. Scientific Reports 7, 9148
| GBF3 transcription factor imparts drought tolerance in Arabidopsis thaliana.Crossref | GoogleScholarGoogle Scholar | 28831141PubMed |
Ravel C, Fiquet S, Boudet J, Dardevet M, Vincent J, Merlino M, Michard R, Martre P (2014) Conserved cis-regulatory modules in promoters of genes encoding wheat high-molecular-weight glutenin subunits. Frontiers in Plant Science 5, 1–17.
| Conserved cis-regulatory modules in promoters of genes encoding wheat high-molecular-weight glutenin subunits.Crossref | GoogleScholarGoogle Scholar |
Roy SC, Shil P (2020) Assessment of genetic heritability in rice breeding lines based on morphological traits and caryopsis ultrastructure. Scientific Reports 10, 7830
| Assessment of genetic heritability in rice breeding lines based on morphological traits and caryopsis ultrastructure.Crossref | GoogleScholarGoogle Scholar | 32385288PubMed |
Schnaithmann F, Pillen K (2013) Detection of exotic QTLs controlling nitrogen stress tolerance among wild barley introgression lines. Euphytica 189, 67–88.
| Detection of exotic QTLs controlling nitrogen stress tolerance among wild barley introgression lines.Crossref | GoogleScholarGoogle Scholar |
Shoemaker CJ, Green R (2012) Translation drives mRNA quality control. Nature Structural & Molecular Biology 19, 594–601.
| Translation drives mRNA quality control.Crossref | GoogleScholarGoogle Scholar |
Song XJ, Kuroha T, Ayano M, Furuta T, Nagai K, Komeda N, Segami S, Miura K, Ogawa D, Kamura T, et al. (2015) Rare allele of a previously unidentified histone H4 acetyltransferase enhances grain weight, yield, and plant biomass in rice. Proceedings of the National Academy of Sciences of the United States of America 112, 76–81.
| Rare allele of a previously unidentified histone H4 acetyltransferase enhances grain weight, yield, and plant biomass in rice.Crossref | GoogleScholarGoogle Scholar | 25535376PubMed |
Srivastava AK, Lu Y, Zinta G, Lang Z, Zhu J-K (2018) UTR-dependent control of gene expression in plants. Trends in Plant Science 23, 248–259.
| UTR-dependent control of gene expression in plants.Crossref | GoogleScholarGoogle Scholar | 29223924PubMed |
Sun S, Wang L, Mao H, Shao L, Li X, Xiao J, Ouyang Y, Zhang Q (2018) A G-protein pathway determines grain size in rice. Nature Communications 9, 851
| A G-protein pathway determines grain size in rice.Crossref | GoogleScholarGoogle Scholar | 29487318PubMed |
Wan XY, Wan JM, Jiang L, Wang JK, Zhai HQ, Weng JF, Wang HL, Lei CL, Wang JL, Zhang X, Cheng ZJ, Guo XP (2006) QTL analysis for rice grain length and fine mapping of an identified QTL with stable and major effects. Theoretical and Applied Genetics 112, 1258–1270.
| QTL analysis for rice grain length and fine mapping of an identified QTL with stable and major effects.Crossref | GoogleScholarGoogle Scholar | 16477428PubMed |
Wang W, Simmonds J, Pan Q, Davidson D, He F, Battal A, Akhunova A, Trick HN, Uauy C, Akhunov E (2018) Gene editing and mutagenesis reveal inter-cultivar differences and additivity in the contribution of TaGW2 homoeologues to grain size and weight in wheat. Theoretical and Applied Genetics 131, 2463–2475.
| Gene editing and mutagenesis reveal inter-cultivar differences and additivity in the contribution of TaGW2 homoeologues to grain size and weight in wheat.Crossref | GoogleScholarGoogle Scholar | 30136108PubMed |
Watt C, Zhou G, McFawn L-A, Chalmers KJ, Li C (2019) Fine mapping of qGL5H, a major grain length locus in barley (Hordeum vulgare L.). Theoretical and Applied Genetics 132, 883–893.
| Fine mapping of qGL5H, a major grain length locus in barley (Hordeum vulgare L.).Crossref | GoogleScholarGoogle Scholar | 30465063PubMed |
Watt C, Zhou G, McFawn L-A, Li C (2020) Fine mapping qGL2H, a major locus controlling grain length in barley (Hordeum vulgare L.). Theoretical and Applied Genetics 133, 2095–2103.
| Fine mapping qGL2H, a major locus controlling grain length in barley (Hordeum vulgare L.).Crossref | GoogleScholarGoogle Scholar | 32193568PubMed |
Wendt T, Holme I, Dockter C, Preuß A, Thomas W, Druka A, Waugh R, Hansson M, Braumann I (2016) HvDep1 is a positive regulator of culm elongation and grain size in barley and impacts yield in an environment-dependent manner. PLoS One 11, e0168924
| HvDep1 is a positive regulator of culm elongation and grain size in barley and impacts yield in an environment-dependent manner.Crossref | GoogleScholarGoogle Scholar | 28005988PubMed |
Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y (2015) The I-TASSER Suite: protein structure and function prediction. Nature Methods 12, 7–8.
| The I-TASSER Suite: protein structure and function prediction.Crossref | GoogleScholarGoogle Scholar | 25549265PubMed |
Ying J-Z, Ma M, Bai C, Huang X-H, Liu J-L, Fan Y-Y, Song X-J (2018) TGW3, a major qtl that negatively modulates grain length and weight in rice. Molecular Plant 11, 750–753.
| TGW3, a major qtl that negatively modulates grain length and weight in rice.Crossref | GoogleScholarGoogle Scholar | 29567450PubMed |
Zhang X, Wang J, Huang J, Lan H, Wang C, Yin C, Wu Y, Tang H, Qian Q, Li J, Zhang H (2012) Rare allele of OsPPKL1 associated with grain length causes extra-large grain and a significant yield increase in rice. Proceedings of the National Academy of Sciences of the United States of America 109, 21534–21539.
| Rare allele of OsPPKL1 associated with grain length causes extra-large grain and a significant yield increase in rice.Crossref | GoogleScholarGoogle Scholar | 23236132PubMed |
Zhang D, Zhang M, Zhou Y, Wang Y, Shen J, Chen H, Zhang L, Lü B, Liang G, Liang J (2019) The rice G protein γ subunit DEP1/qPE9-1 positively regulates grain-filling process by increasing auxin and cytokinin content in rice grains. Rice 12, 91–102.
| The rice G protein γ subunit DEP1/qPE9-1 positively regulates grain-filling process by increasing auxin and cytokinin content in rice grains.Crossref | GoogleScholarGoogle Scholar | 31844998PubMed |