Host-induced silencing of a nematode chitin synthase gene decreases abundance of rhizosphere fungal community while enhancing Heterodera glycines resistance of soybean
Shuan Tian A # , Xue Shi A # , Baoyuan Qu B , Houxiang Kang A , Wenkun Huang A , Huan Peng A , Deliang Peng A , Jiajun Wang C , Shiming Liu A * and Lingan Kong A *
A , Jiajun Wang C , Shiming Liu A * and Lingan Kong A *
A State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing 100193, China.
B State Key Laboratory of Plant Genomics, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing 100101, China.
C Institute of Soybean Research, Heilongjiang Academy of Agricultural Sciences, Harbin 150086, China.
Handling Editor: Marta Santalla
Crop & Pasture Science 73(10) 1156-1167 https://doi.org/10.1071/CP22030
Submitted: 14 December 2021 Accepted: 10 March 2022 Published: 23 April 2022
© 2022 The Author(s) (or their employer(s)). Published by CSIRO Publishing. This is an open access article distributed under the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License (CC BY-NC-ND)
Abstract
Context: A transgenic variety of soybean (Glycine max (L.) Merr.), H57, has been developed from wild-type variety Jack, with host-induced gene silencing of a chitin synthase gene (CHS) in soybean cyst nematode (SCN, Heterodera glycines Ichinohe), a devastating pathogen in soybean. H57 needs to be characterised for suitability to manage SCN, especially because rhizosphere microbial communities may be sensitive to genetically modified crops.
Aims: We aimed to evaluate the SCN resistance of H57 at the T7 generation, and analyse the impact on the rhizosphere microbial community of planting H57 into SCN-infected soil.
Methods: Infection with SCN was assessed at 60 days after planting of H57 and Jack into SCN-infected soil by examining recovered cysts from rhizosphere soil and comparing with an infected bulk soil control. For analysis of rhizosphere microbial communities (bacterial and fungal), 16S and ITS amplicons were identified by high-throughput sequencing, and bioinformatic analysis was used to define operational taxonomic units. Alpha diversity, using five indexes, and relative abundance were determined.
Key results: Soybean H57 showed significantly enhanced and heritable resistance to SCN compared with Jack. The diversity and richness (abundance) of the bacterial community of H57 and Jack were significantly and similarly increased relative to the bulk soil. The fungal community of H57 had considerably lower abundance than both other treatments, and lower diversity than the bulk soil. The relative abundance of only two bacterial phyla (Acidobacteria and Actinobacteria) and one fungal phylum (Glomeromycota), and three bacterial genera (Candidatus_Solibacter, Candidatus_Udaeobacter and Bryobacter) and one fungal genus (Aspergillus), differed significantly between rhizosphere soils of H57 and Jack.
Conclusions: Host-induced gene silencing of SCN-CHS substantially and heritably enhanced SCN resistance in soybean, did not significantly alter the rhizosphere bacterial community, but greatly suppressed the abundance of the rhizosphere fungal community, which was likely associated with boosted SCN resistance.
Implications: This study established a basis for interaction research between soybean with SCN-CHS host-induced gene silencing and the rhizosphere microbial community, and for potentially planting soybean H57 to manage SCN.
Keywords: abundance, chitin synthase, diversity, host-induced silencing of gene, rhizosphere microbial community, SCN resistance, soybean, transgenic soybean H57.
Introduction
Soybean (Glycine max (L.) Merr.) is a major crop globally, as an important source of sustainable supply of protein and oil. A steady increase in soybean production is required to meet growing world food and energy demands. Soybean cyst nematode (SCN, Heterodera glycines Ichinohe) is one of the most economically damaging pathogens in soybean worldwide, significantly constraining soybean production and causing huge yield losses annually (Peng et al. 2021). SCN secretes a series of effectors mainly through a spear-like feeding structure called a stylet, penetrating host root cell walls and inducing the formation of complex feeding sites (syncytia) near the vascular cylinder, from which SCN obtains nutrition. Aboveground symptoms are not always visible after infection by SCN, meaning that infection is usually recognised at later stages of infection when significant damage has already occurred; the nematode is difficult to eradicate, and damage is often devastating (Niblack et al. 2006). Applications of fertilisers and pesticides provide some mitigation, but the use of improved cultivars with underlying resistance to pathogens is the most attractive option for farmers in soybean production (Kim et al. 2016; Shaibu et al. 2020).
Host-induced gene silencing (HIGS) has shown potential for controlling various pathogens, through generation of transgenic plants carrying hairpin constructs against the target genes in the harmful organisms (Mao et al. 2007; Zhang et al. 2016). For instance, HIGS targeting of fungal genes is reported to be an efficient strategy for controlling the fungal diseases caused by Blumeria graminis, Fusarium graminearum and Verticillium dahlia (Nowara et al. 2010; Cheng et al. 2015; Zhang et al. 2016). A high level of accumulated small interference RNAs (siRNAs) played an important role in the defence of HIGS cotton plants against pathogenic V. dahlia (Zhang et al. 2016). However, there are only a few preliminary studies on the application of HIGS against plant parasitic nematodes. In eggplant, HIGS targeting three nematode effector genes (Mi-msp-1, Mi-msp-18 and Mi-msp-20), whose transcripts were accumulated in subventral pharyngeal gland cells of Meloidogyne incognita, conferred resistance to the nematode (Shivakumara et al. 2017; Chaudhary et al. 2019). In hairy roots of soybean, HIGS targeting six reproduction or fitness-related genes (Cpn-1, Y25, Prp-17, J15, J20 and J23), showed a significant suppression of SCN populations; however, HIGS was confined to transient composite soybeans, and not heritable (Li et al. 2010; Tian et al. 2016). To date, HIGS soybean lines with broad-spectrum and durable resistance to H. glycines have not been reported.
Chitin is a linear polymer of b-(1,4)-linked N-acetylglucosamine (GlcNAc) synthesised by chitin synthase (Chs) that is present in fungi and nematodes but absent in plants and vertebrate animals (Kong et al. 2012). Therefore, Chs is widely considered an important target for designing new fungicides and developing novel resistant crop varieties via HIGS (Kong et al. 2012; Cheng et al. 2015). Our previous studies demonstrated three CHS genes (MoCHS1, MoCHS6 and MoCHS7) in a model phytopathogenic fungus Magnaporthe oryzae as potential targets for developing novel fungicides (Kong et al. 2012). HIGS wheat lines targeting a CHS gene (Chs3b) in F. graminearum showed durable resistance to Fusarium head blight and seedling blight, even the HIGS-T5 generation showing strong resistance (Cheng et al. 2015).
Interactions between plants and rhizosphere microorganisms are vital in carbon sequestration, ecosystem functioning and nutrient cycling in natural ecosystems as well as agricultural and forest systems (Singh et al. 2004), and the rhizosphere microbial community is often influenced by plant genotype and environmental conditions (Filion 2008). Plants interact with soil microorganisms, thereby affecting rhizosphere microbial community dynamics (Wu et al. 2014). Diversity changes in rhizosphere microbial communities may influence bio- and geo-chemical processes in soil ecology (Chauhan et al. 2014; Kostov et al. 2014). Rhizosphere microbial communities may be very sensitive to genetically modified (transgenic) crops (Gao et al. 2015). Therefore, the structure/diversity and abundance of rhizosphere microbial communities are often used as an early and sensitive indicator for assessing the effects of transgenic crops on soil ecology (Liang et al. 2018).
We recently cloned a soybean cyst nematode chitin synthase gene (SCN-CHS), and generated transgenic soybean lines with HIGS of SCN-CHS, using soybean variety Jack as the wild-type. The T2 and T6 generations of the three homozygous SCN-CHS HIGS lines that we obtained all showed enhanced resistance to SCN (Kong et al. 2022). In the present study, the main aims were (i) to evaluate the resistance in the T7 generation of one of the transgenic soybean lines, H57, to SCN (H. glycines) and its inheritance; and (ii) to analyse whether and what in detail HIGS of SCN-CHS alters the rhizosphere microbial community, by using high-throughput sequencing and analyses of bacterial 16S rRNA genes and fungal internal transcribed spacer (ITS) sequences. The results obtained will provide a basis for further research on the interactions between SCN-CHS HIGS soybean and the rhizosphere microbial community, and for extensive planting of SCN-CHS HIGS soybean for effective management of SCN.
Materials and methods
Soybean lines
The sole CHS gene with 3984 bp (GenBank Acc. No.: OK149168) was cloned from H. glycines HG Type 1.2.3.5.7 and designated as SCN-CHS. SCN-Chs contains a typical chitin synthase catalytic domain (Chs) and seven transmembrane domains (TMs). A 420 bp cDNA fragment of the SCN-Chs core region positioned at 1936–2355 bp was cloned and induced into soybean variety Jack to generate HIGS transgenic soybeans by Dabeinong Science and Technology Group Co. Ltd., China (Kong et al. 2022). H57 was one of the homozygous transgenic soybean lines identified. H57 was raised to the T7 generation by our laboratory in the greenhouse of Langfang Experimental Base of Institute of Plant Protection, Chinese Academy of Agricultural Sciences, for the following experiments. Jack was used as the control in this study.
Soybean growth, SCN life cycle and soil sampling
Seeds of the T7 generation of H57 and Jack (as control) were planted in six pots filled with SCN-infected soil collected from a field that was severely infected with SCN HG Type 0, with ∼6000 eggs (40 cysts)/100 g dry soil, in Jiayin County, Yichun City, Heilongjiang Province, China. Plants were grown in a greenhouse under conditions of 16 h light at 28°C and 8 h dark at 26°C. A plot with six pots comprising the same soil with no planting was used as a bulk soil control.
After SCN eggs develop into first-stage juveniles (J1s) and J2s in the eggs, the parasitic J2s (par-J2s) hatch from the eggs and penetrate the soybean roots, migrating near to the vascular cylinder to induce the formation of syncytia. The par-J2s finish their life cycle through successive J3, J4 and adult (female and male) stages by obtaining nutrition from syncytia. Each female can fill up with ≥200 eggs after mating with males on the root surface, and finally, the egg-filled females die and their body wall hardens to form a tough cyst around the eggs.
At 60 days after planting, soil samples were collected from the rhizosphere of soybean-planted plots and from the bulk soil plot, as described previously (Jin et al. 2021). Six soybean seedlings were collected from H57 and Jack plots. Roots were shaken free of soil, and rhizosphere soil fractions were brushed and pooled into one sample (one replicate). Six replicates of soil samples were collected for each treatment (bulk soil, and rhizosphere soil of H57 or Jack); in total, 18 soil samples were obtained and used for SCN-infection phenotype evaluation and sequencing.
SCN-infection phenotype of soybeans
Cysts were collected from the rhizosphere soil samples of each treatment by suspending the soil in water and filtering the samples through nested sieves of 710 μm and 250 μm apertures. The recovered cysts were counted under an SZ61 stereo microscope (Olympus, Tokyo, Japan). The full shape of cysts was captured to measure their length and width. Ten plump cysts of similar size were randomly selected from each treatment. They were placed on a slide with a drop of water and gently punctured with tweezers, and the number of eggs in each cyst was counted under the stereo microscope.
High-throughput sequencing
Genomic DNA was extracted from each soil sample by using a Power Soil DNA Isolation Kit (MO BIO Laboratories, Carlsbad, CA, USA) according to the protocol of the manufacturer. Extracted DNA was quantified by ratios of 260 nm/280 nm and 260 nm/230 nm. The V3–V4 region of the bacterial 16S rRNA gene was amplified using the common primer pair (forward primer 5′-ACTCCTACGGGAGGCAGCA-3′; reverse primer 5′-GGACTACHVGGGTWTCTAAT-3′) and the fungal ITS1 region using the primer pair (forward primer 5′-CTTGGTCATTTAGAGGAAGTAA-3′; reverse primer 5′-GCTGCGTTCTTCATCGATGC-3′) combined with adapter sequences and barcode sequences. PCR amplification was conducted in a total volume of 50 μL, which contained 10 μL buffer, 0.2 μL Q5 High-Fidelity DNA Polymerase, 10 μL High GC Enhancer (New England Biolabs, Ipswich, MA, USA), 1 μL dNTP, 10 μM each primer and 60 ng genomic DNA. Thermal cycling conditions were as follows: an initial denaturation at 95°C for 5 min, followed by 15 cycles of 95°C for 1 min, 50°C for 1 min and 72°C for 1 min, with a final extension at 72°C for 7 min. The PCR products from the first-step PCR were purified using VAHTS DNA Clean Beads (Vazyme Biotech, Nanjing, China). A second-round PCR was then performed in a 40-μL reaction volume containing 20 μL 2× Phusion HF MM (Thermo Fisher Scientific, Waltham, MA, USA), 8 μL ddH2O, 10 μM each primer and 10 μL PCR products from the first step. Thermal cycling conditions were as follows: an initial denaturation at 98°C for 30 s, followed by 10 cycles of 98°C for 10 s, 65°C for 30 s and 72°C for 30 s, with a final extension at 72°C for 5 min. Finally, all PCR products were quantified using Quant-iT dsDNA HS Reagent (Thermo Fisher Scientific) and pooled. High-throughput sequencing analysis of bacterial rRNA genes and the fungal ITS region was conducted on the purified and pooled sample using the HiSeq 2500 platform (Illumina, San Diego, CA, USA) (2× 250 paired ends) at Biomarker Technologies, Beijing, China.
Data analyses
The raw tags were generated by FLASH software (version 1.2.7; Center for Computational Biology, Washington, DC, USA) based on the overlap of paired-end reads, and filtered by Trimmomatic software (version 0.33; Bolger et al. 2014) to obtain the clean tags. Effective tags were acquired by removing chimera sequences from the clean tags using UCHIME software (version 4.2; Edgar 2017), and then clustered as operational taxonomic units (OTUs) using QIIME software (ver. 1.8.0; Caporaso et al. 2010). OTUs were annotated according to Silva database (https://www.arb-silva.de/), and different levels of classification tables were also generated.
Alpha diversity was applied to reflect the richness and diversity of species through five indexes: observed species, Shannon, Simpson, Chao1 and Ace. Community structure was analysed using principal components analysis (PCA). Analyses were performed using BMKCloud (http://www.biocloud.net) at the OTU level. The coordinate graphs were redrawn in RStudio (ver. 3.5.3; RStudio, Boston, MA, USA) based on the PCA coordinates and explanatory information. The relative abundance of phyla was analysed by BMKCloud to show the top 10 bacterial and fungal phyla based on their overall abundance in the bulk soil, and the rhizosphere soils of Jack and H57. Analysis of variance was performed using SPSS Statistics 22 (IBM, Armonk, NY, USA). Duncan’s Multiple-Range tests were performed to test for significant difference among treatments. All the statistical tests were performed using P = 0.05 as the level of significance.
Results
SCN-CHS HIGS soybean H57 showed enhanced resistance to H. glycines
Soybean H57 is one of three generated homozygous SCN-CHS HIGS lines whose T2 and T6 generations show enhanced SCN resistance over the wild-type variety Jack, with significant silencing (suppression) of SCN-CHS in nematodes after infection (Kong et al. 2022). Here, the T7 generation of SCN-CHS HIGS soybean H57 was evaluated for its heritable resistance to SCN in the greenhouse. Relative to Jack, the number of H. glycines cysts (Fig. 1a), number of eggs per cyst (Fig. 1b) and diameter of cysts (Fig. 1c) in H57 were significantly (P < 0.05) reduced by 76.7%, 21.0% and 6.7%, respectively, at 60 days post planting. Therefore, SCN-CHS HIGS soybean H57 showed enhanced resistance to H. glycines over the variety from which it was generated.
Sequencing summary
High-throughput sequencing of 18 samples yielded 1 440 762 raw reads of the 16S rRNA gene. After quality filtering, 1 434 149 clean reads remained. In total, 1 394 746 effective reads were obtained for further OTU clustering. Sequence clustering yielded 2034 OTUs, which were assigned to 36 phyla, 94 classes, 209 orders, 342 families, 586 genera and 636 species. On the other hand, 1 411 108 raw fungal ITS gene sequences were obtained after sequencing of 18 samples. After quality filtering, 1 390 318 clean reads remained, with 1 382 619 effective reads acquired for further OTU clustering. Sequence clustering yielded 759 OTUs, which were assigned to 11 phyla, 29 classes, 65 orders, 119 families 215 genera and 250 species. All of the rarefaction curves tended to approach a plateau, indicating that the numbers of sequences acquired were sufficient to describe the bacterial and fungal diversity and abundance within these samples (Fig. 2).
Difference in diversity and richness of microbial community between rhizosphere soil of SCN-CHS HIGS soybean H57 and wild-type Jack
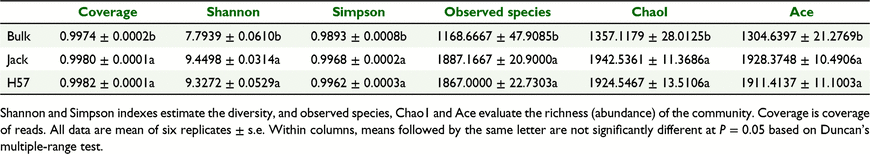
Alpha diversity of the microbial community of rhizosphere soil of H57 and Jack, and the bulk soil, is represented by five indexes; Shannon and Simpson were used to estimate the diversity, whereas observed species, Chao1 and Ace were employed to evaluate the richness (abundance), of the rhizosphere bacterial (Table 1) and fungal (Table 2) communities. The coverage of clean reads was >99% for all soil samples, indicating that depth of sequencing was sufficient to represent the majority of the microbiota in the soybean rhizosphere soil environment. Therefore, the obtained sequencing data can be used for reliably analysing the diversity and abundance of the microbial community.
The results indicate that planting soybeans (H57 or Jack) significantly increased both diversity and abundance of the rhizosphere bacterial community relative to the bulk soil; however, comparing the rhizosphere soils of H57 and Jack, both diversity and abundance of the bacterial communities were similar (Table 1). Therefore, HIGS of SCN-CHS did not significantly impact either the diversity or the abundance of rhizosphere bacterial community. Regarding the fungal community, planting Jack did not significantly change the diversity or abundance of the soil fungal community relative to bulk soil. On the other hand, planting H57 significantly reduced both the diversity and abundance (Chao1 and Ace, but not observed species) of the soil fungal community relative to bulk soil. Fungal diversity was not significantly different between plantings of H57 and Jack; however, abundance (observed species, Chao1 and Ace) of the rhizosphere fungal community was significantly lower with H57 than with Jack (Table 2). These results suggest that HIGS of SCN-CHS significantly decreased the abundance of rhizosphere fungal community.

|
The PCA indicated that the bacterial community structure/diversity was similar in the rhizosphere soil of Jack and H57, and significantly different in the bulk soil (Fig. 3a). The structure/diversity of the rhizosphere fungal community of H57 was different from that in the bulk soil, and that of the rhizosphere soil of Jack was partially similar to that in the bulk soil (Fig. 3b). A Venn diagram can be used to display the numbers of common and unique features of OTUs, and intuitively shows the coincidence of features, between samples. The number of common features of the bacterial community between the rhizosphere soil samples of H57 and Jack was 2012, with a similarity of 99.1% (Fig. 4a), indicating extreme similarity in the components/diversity of the bacterial community in the rhizosphere soil of H57 and Jack. The number of common features of the fungal community between the rhizosphere soil samples of H57 and Jack was 573, with a similarity of only 77.3% (Fig. 4b), suggesting partial differences in the diversity of the fungal community in the rhizosphere soil of H57 and Jack. All of these results further validate that planting of H57 and Jack similarly affected the structure/diversity of the rhizosphere bacterial community, but had some different impacts on the rhizosphere fungal community.
Difference in relative abundance of microbial community at phylum level between rhizosphere soil of SCN-CHS HIGS soybean H57 and wild-type Jack
Analyses were conducted for the rhizosphere microbial community at the phylum level, with the relative abundance of the top 10 phyla in bulk soil and rhizosphere soil of Jack and H57 presented in Fig. 5a and b, for bacteria and fungi, respectively. The relative abundance of the top 10 bacterial phyla except Chloroflexi in the bulk soil was significantly (P < 0.05) different from that in the rhizosphere soil of H57 and Jack (Fig. 5c). However, the relative abundance of only one of the top 10 fungal phyla was significantly (P < 0.05) altered in the rhizosphere soil of H57 compared with Jack (Fig. 5b). In all soil samples, Proteobacteria and Acidobacteria were the two most dominant bacterial phyla, accounting for >61% of the reads (Fig. 5a). Ascomycota and Basidiomycota were the two main fungal phyla, occupying >57% of the reads (Fig. 5b). Furthermore, after planting H57, the relative abundance of only two bacterial phyla, Acidobacteria and Actinobacteria, was significantly (P < 0.05) different from that in the rhizosphere soil of Jack (Fig. 5c); abundance of Acidobacteria was higher, and that of Actinobacteria was much lower, in the rhizosphere soil of H57. Similarly, the relative abundance of only one fungal phylum, Glomeromycota, showed significant difference between the rhizosphere soils of H57 and Jack (Fig. 5d).
Difference in relative abundance of microbial community at genus level between rhizosphere soil of SCN-CHS HIGS soybean H57 and wild-type Jack
At the genus level, the relative abundance of the bacteria Candidatus_Solibacter, Candidatus_Udaeobacter and Bryobacter in the rhizosphere soil of H57 was different from that of Jack (Fig. 6a, Table 3). Meanwhile, the relative abundance of fungal genera Arcoplius, Arxotrichum, Chaetomidium, Chaetomium, Chrysanthotrichum, Condenascus, Mycofalcella and Penicillium was significantly changed in the rhizosphere soil of Jack, whereas that of fungal Aspergillus was not significantly altered, compared with the bulk soil (Fig. 6b, Table 4). However, after planting H57, the relative abundance of rhizosphere Aspergillus was considerably lower than after planting Jack. Aspergillus is the sole genus of the top 20 fungal genera presenting significant (P < 0.05) abundance difference between the rhizosphere soils of Jack and H57 (Fig. 6b, Table 4).

|

|
Discussion
In this study, a 76.7% decrease of H. glycines cyst numbers and significant reductions in numbers of eggs per cyst and cyst diameters in the plots with T7 generation SCN-CHS HIGS transgenic soybean H57 (Fig. 1) indicate its significantly enhanced resistance. In addition to enhanced SCN resistance phenotype results for T2 and T6 generations of SCN-CHS HIGS soybean lines (Kong et al. 2022), this clearly indicates that the enhanced resistance to SCN is highly heritable in HIGS soybean H57, establishing a strong basis for planting H57 for effective management of SCN. Meanwhile, planting SCN-CHS HIGS soybean H57 and the wild-type Jack almost equally significantly enhanced the diversity and richness (abundance) of the rhizosphere bacterial community (Table 1, Fig. 3a), whereas planting H57 rather than Jack greatly decreased the abundance of the rhizosphere fungal community, and planting soybean H57 significantly reduced its diversity compared with bulk soil (Table 2), suggesting that HIGS of SCN-CHS mainly altered the abundance of the rhizosphere fungal community.
After planting H57, only two rhizosphere bacterial phyla, Acidobacteria and Actinobacteria, were significantly altered compared with planting Jack; relative abundance of Acidobacteria was significantly increased, and relative abundance of Actinobacteria was greatly decreased. Acidobacteria was one of the two most predominant bacterial phyla, and was much more abundant than Actinobacteria in all soil samples (Fig. 5a, c). Thus, Acidobacteria should contribute more to differences in the bacterial community between the rhizosphere soils of H57 and Jack. Furthermore, at the genus level, the abundance of Bryobacter, which belongs to Acidobcateria phylum, was also much lower in the rhizosphere soil of H57 than Jack (Fig. 6a, Table 3). These results indicate that HIGS of SCN-CHS stimulates the growth of Acidobacteria while suppresses Bryobacter. The ubiquity of Acidobacteria in soil indicates their importance in the functioning of the soil ecosystem (Ward et al. 2009); for example, they might be responsible for degrading polysaccharides of plants and fungi in the ecosystems of acidic coniferous forests (Lladó et al. 2016). Genomic and metagenomic data predict a number of ecologically relevant capabilities of some Acidobacteria, which include the ability to use nitrite as a nitrogen source, respond to soil macro- and micro-nutrients and soil acidity, express multiple active transporters, degrade gellan gum, and produce exopolysaccharide (Kielak et al. 2016). Therefore, we propose that higher abundance of Acidobacteria in the rhizosphere soil of H57 might degrade more polysaccharides of fungal cell walls, resulting in suppression of the rhizosphere fungal community compared with the rhizosphere soil of Jack.
At the fungal phylum level, only Glomeromycota was significantly different between the rhizosphere soils of H57 and Jack. Fungi in the phylum Glomeromycota include the arbuscular mycorrhizal fungi (AMF), which are are symbiotic with plant roots, and therefore the most common fungi in the terrestrial ecosystem (Lee et al. 2018). AMF are also associated with vascular plants and thalloid bryophytes (Brundrett and Tedersoo 2018). In addition, the species richness of sporulating AMF is high in Roraima savannas of Brazil, indicating that AMF may play a key role in plant nutrition and soil structure improvement in the presence of poor soil nutrients or other harsh conditions (Stürmer et al. 2018). However, in our study, the relative abundance of Glomeromycota in each soil sample was ≤1% (Fig. 5b, d). Thus, Glomeromycota is a minor fungal phylum in all soil samples in this study. Nonetheless, the functions of fungal Glomeromycota in SCN-CHS HIGS soybean H57 and the rhizosphere microbial community interaction system are worthy of further investigation.
At the fungal genus level, Aspergillus was the sole genus that showed significant difference between the rhizosphere soil of H57 and Jack (Fig. 6b, Table 4). Some Aspergillus spp. such as A. welwitschiae (Liu et al. 2019) and A. niger NBC001 (Jin et al. 2019) have been isolated and can be used as biocontrol reagents for effectively controlling root-knot nematode (Meloidogyne graminicola) (Shemshura et al. 2016; Liu et al. 2019), and SCN (Jin et al. 2019, 2021), but most Aspergillus spp. are pathogenic. The much lower abundance of Aspergillus in the rhizosphere soil of H57 than Jack (Fig. 6b, Table 4) indicates that HIGS of SCN-CHS inhibits rhizosphere Aspergillus. Our recent study demonstrated that HIGS of SCN-CHS could significantly suppress soybean infection by Fusarium oxysporum, which causes Fusarium wilt disease in soybean (Kong et al. 2022). The present results suggest that HIGS of SCN-CHS can suppress the relative abundance of pathogenic fungi, leading to decreased abundance of the rhizosphere fungal community (Table 2) while enhancing the resistance of soybean to SCN (Fig. 1). The significantly increased relative abundance of the bacterial phylum Acidobacteria under planting of SCN-CHS HIGS H57 may increase degradation of polysaccharides in fungal cell walls and result in reduced abundance of the rhizosphere fungal community. Therefore, we hypothesise that HIGS of SCN-CHS compromises the abundance of the rhizosphere fungal community, likely through stimulation of the diversity and abundance of the rhizosphere bacterial community to enhance the resistance of soybean to SCN (H. glycines), but the mechanisms require further study.
Data availability
All the data are included in this manuscript.
Conflicts of interest
The authors declare no conflicts of interest.
Declaration of funding
This study was funded by grants from the National Natural Science Foundation of China (31872924, 31972248), the National Key Research and Development Program of China (2018YFD0201002), and the State Key Laboratory for Biology of Plant Diseases and Insect Pests (SKLOF201613).
Acknowledgements
We thank Dabeinong Science and Technology Group Co. Ltd, China for developing the HIGS transgenic H57, and Biomarker Technologies Corporation (Beijing) for conducting 16S/ITS high-throughput sequencing.
References
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120.| Trimmomatic: a flexible trimmer for Illumina sequence data.Crossref | GoogleScholarGoogle Scholar | 24695404PubMed |
Brundrett MC, Tedersoo L (2018) Evolutionary history of mycorrhizal symbioses and global host plant diversity. New Phytologist 220, 1108–1115.
| Evolutionary history of mycorrhizal symbioses and global host plant diversity.Crossref | GoogleScholarGoogle Scholar |
Caporaso JG, Kuczynski J, Stombaugh J, et al. (2010) QIIME allows analysis of high-throughput community sequencing data. Nature Methods 7, 335–336.
| QIIME allows analysis of high-throughput community sequencing data.Crossref | GoogleScholarGoogle Scholar | 20383131PubMed |
Chaudhary S, Dutta TK, Tyagi N, Shivakumara TN, Papolu PK, Chobhe KA, Rao U (2019) Host-induced silencing of Mi-msp-1 confers resistance to root-knot nematode Meloidogyne incognita in eggplant. Transgenic Research 28, 327–340.
| Host-induced silencing of Mi-msp-1 confers resistance to root-knot nematode Meloidogyne incognita in eggplant.Crossref | GoogleScholarGoogle Scholar | 30955133PubMed |
Chauhan A, Smartt A, Wang J, et al. (2014) Integrated metagenomics and metatranscriptomics analyses of root-associated soil from transgenic switchgrass. Genome Announcements 2, e00777-14
| Integrated metagenomics and metatranscriptomics analyses of root-associated soil from transgenic switchgrass.Crossref | GoogleScholarGoogle Scholar | 25125642PubMed |
Cheng W, Song XS, Li HP, Cao LH, Sun K, Qiu XL, Xu YB, Yang P, Huang T, Zhang JB, Qu B, Liao YC (2015) Host-induced gene silencing of an essential chitin synthase gene confers durable resistance to Fusarium head blight and seedling blight in wheat. Plant Biotechnology Journal 13, 1335–1345.
| Host-induced gene silencing of an essential chitin synthase gene confers durable resistance to Fusarium head blight and seedling blight in wheat.Crossref | GoogleScholarGoogle Scholar | 25735638PubMed |
Edgar R (2017) SEARCH_16S: a new algorithm for identifying 16S ribosomal RNA genes in contigs and chromosomes. BioRxiv
| SEARCH_16S: a new algorithm for identifying 16S ribosomal RNA genes in contigs and chromosomes.Crossref | GoogleScholarGoogle Scholar |
Filion M (2008) Do transgenic plants affect rhizobacteria populations? Microbial Biotechnology 1, 463–475.
| Do transgenic plants affect rhizobacteria populations?Crossref | GoogleScholarGoogle Scholar | 21261867PubMed |
Gao N, Shen W, Lin X, Shi W (2015) Influence of transgenic ath-miR399d tomato lines on microbial community and diversity in rhizosphere soil. Soil Science and Plant Nutrition 61, 259–268.
| Influence of transgenic ath-miR399d tomato lines on microbial community and diversity in rhizosphere soil.Crossref | GoogleScholarGoogle Scholar |
Jin N, Liu SM, Peng H, Huang WK, Kong LA, Wu YH, Chen Y, Ge FY, Jian H, Peng DL (2019) Isolation and characterization of Aspergillus niger NBC001 underlying suppression against Heterodera glycines. Scientific Reports 9, 591
| Isolation and characterization of Aspergillus niger NBC001 underlying suppression against Heterodera glycines.Crossref | GoogleScholarGoogle Scholar | 30679719PubMed |
Jin N, Liu SM, Peng H, Huang WK, Kong LA, Peng DL (2021) Effect of Aspergillus niger NBC001 on the soybean rhizosphere microbial community in a soybean cyst nematode-infested field. Journal of Integrative Agriculture 20, 3230–3239.
| Effect of Aspergillus niger NBC001 on the soybean rhizosphere microbial community in a soybean cyst nematode-infested field.Crossref | GoogleScholarGoogle Scholar |
Kielak AM, Barreto CC, Kowalchuk GA, van Veen JA, Kuramae EE (2016) The ecology of Acidobacteria: moving beyond genes and genomics. Frontiers in Microbiology 7, 744
| The ecology of Acidobacteria: moving beyond genes and genomics.Crossref | GoogleScholarGoogle Scholar | 27303369PubMed |
Kim KS, Vuong TD, Qiu D, Robbins RT, Grover Shannon J, Li Z, Nguyen HT (2016) Advancements in breeding, genetics, and genomics for resistance to three nematode species in soybean. Theoretical and Applied Genetics 129, 2295–2311.
| Advancements in breeding, genetics, and genomics for resistance to three nematode species in soybean.Crossref | GoogleScholarGoogle Scholar | 27796432PubMed |
Kong LA, Yang J, Li GT, Qi LL, Zhang YJ, Wang CF, Zhao WS, Xu JR, Peng YL (2012) Different chitin synthase genes are required for various developmental and plant infection processes in the rice blast fungus Magnaporthe oryzae. PLoS Pathogens 8, e1002526
| Different chitin synthase genes are required for various developmental and plant infection processes in the rice blast fungus Magnaporthe oryzae.Crossref | GoogleScholarGoogle Scholar | 22346755PubMed |
Kong L, Shi X, Chen D, Yang N, Yin C, Yang J, Wang G, Huang G, Peng H, Peng D, Liu S (2022) Host-induced silencing of a nematode chitin synthase gene enhances resistance of soybeans to both pathogenic Heterodera glycines and Fusarium oxysporum. Plant Biotechnology Journal
| Host-induced silencing of a nematode chitin synthase gene enhances resistance of soybeans to both pathogenic Heterodera glycines and Fusarium oxysporum.Crossref | GoogleScholarGoogle Scholar | 35301818PubMed |
Kostov K, Krogh PH, Damgaard CF (2014) Are soil microbial endpoints changed by Bt crops compared with conventional crops? A systematic review protocol. Environmental Evidence 3, 11
| Are soil microbial endpoints changed by Bt crops compared with conventional crops? A systematic review protocol.Crossref | GoogleScholarGoogle Scholar |
Lee SI, Lee KJ, Chun HH, Ha S, Gwak HJ, Kim HM, Lee JH, Choi HJ, Kim HH, Shin TS, Park HW, Kim JC (2018) Process development of oxalic acid production in submerged culture of Aspergillus niger F22 and its biocontrol efficacy against the root-knot nematode Meloidogyne incognita. Bioprocess and Biosystems Engineering 41, 345–352.
| Process development of oxalic acid production in submerged culture of Aspergillus niger F22 and its biocontrol efficacy against the root-knot nematode Meloidogyne incognita.Crossref | GoogleScholarGoogle Scholar | 29150701PubMed |
Li J, Todd TC, Oakley TR, Lee J, Trick HN (2010) Host-derived suppression of nematode reproductive and fitness genes decreases fecundity of Heterodera glycines Ichinohe. Planta 232, 775–785.
| Host-derived suppression of nematode reproductive and fitness genes decreases fecundity of Heterodera glycines Ichinohe.Crossref | GoogleScholarGoogle Scholar | 20582434PubMed |
Liang J, Jiao Y, Luan Y, Sun S, Wu C, Wu H, Zhang M, Zhang H, Zheng X, Zhang Z (2018) A 2-year field trial reveals no significant effects of GM high-methionine soybean on the rhizosphere bacterial communities. World Journal of Microbiology and Biotechnology 34, 113
| A 2-year field trial reveals no significant effects of GM high-methionine soybean on the rhizosphere bacterial communities.Crossref | GoogleScholarGoogle Scholar | 29987404PubMed |
Liu Y, Ding Z, Peng DL, Liu SM, Kong LA, Peng H, Xiang C, Li ZC, Huang WK (2019) Evaluation of the biocontrol potential of Aspergillus welwitschiae against the root-knot nematode Meloidogyne graminicola in rice (Oryza sativa L.). Journal of Integrative Agriculture 18, 2561–2570.
| Evaluation of the biocontrol potential of Aspergillus welwitschiae against the root-knot nematode Meloidogyne graminicola in rice (Oryza sativa L.).Crossref | GoogleScholarGoogle Scholar |
Lladó S, Žifčáková L, Větrovský T, Eichlerová I, Baldrian P (2016) Functional screening of abundant bacteria from acidic forest soil indicates the metabolic potential of Acidobacteria subdivision 1 for polysaccharide decomposition. Biology and Fertility of Soils 52, 251–260.
| Functional screening of abundant bacteria from acidic forest soil indicates the metabolic potential of Acidobacteria subdivision 1 for polysaccharide decomposition.Crossref | GoogleScholarGoogle Scholar |
Mao YB, Cai WJ, Wang JW, Hong GJ, Tao XY, Wang LJ, Huang YP, Chen XY (2007) Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nature Biotechnology 25, 1307–1313.
| Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol.Crossref | GoogleScholarGoogle Scholar | 17982444PubMed |
Niblack TL, Lambert KN, Tylka GL (2006) A model plant pathogen from the kingdom Animalia: Heterodera glycines, the soybean cyst nematode. Annual Review of Phytopathology 44, 283–303.
| A model plant pathogen from the kingdom Animalia: Heterodera glycines, the soybean cyst nematode.Crossref | GoogleScholarGoogle Scholar | 16704359PubMed |
Nowara D, Gay A, Lacomme C, Shaw J, Ridout C, Douchkov D, Hensel G, Kumlehn J, Schweizer P (2010) HIGS: host-induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis. The Plant Cell 22, 3130–3141.
| HIGS: host-induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis.Crossref | GoogleScholarGoogle Scholar | 20884801PubMed |
Peng D, Jiang R, Peng H, Liu S (2021) Soybean cyst nematodes: a destructive threat to soybean production in China. Phytopathology Research 3, 19
| Soybean cyst nematodes: a destructive threat to soybean production in China.Crossref | GoogleScholarGoogle Scholar |
Shaibu AS, Li B, Zhang S, Sun J (2020) Soybean cyst nematode-resistance: gene identification and breeding strategies. The Crop Journal 8, 892–904.
| Soybean cyst nematode-resistance: gene identification and breeding strategies.Crossref | GoogleScholarGoogle Scholar |
Shemshura ON, Bekmakhanova NE, Mazunina MN, Meyer SLF, Rice CP, Masler EP (2016) Isolation and identification of nematode-antagonistic compounds from the fungus Aspergillus candidus. FEMS Microbiology Letters 363, fnw026
| Isolation and identification of nematode-antagonistic compounds from the fungus Aspergillus candidus.Crossref | GoogleScholarGoogle Scholar | 26850440PubMed |
Shivakumara TN, Chaudhary S, Kamaraju D, Dutta TK, Papolu PK, Banakar P, Sreevathsa R, Singh B, Manjaiah KM, Rao U (2017) Host-induced silencing of two pharyngeal gland genes conferred transcriptional alteration of cell wall-modifying enzymes of Meloidogyne incognita vis-à-vis perturbed nematode infectivity in eggplant. Frontiers in Plant Science 8, 473
| Host-induced silencing of two pharyngeal gland genes conferred transcriptional alteration of cell wall-modifying enzymes of Meloidogyne incognita vis-à-vis perturbed nematode infectivity in eggplant.Crossref | GoogleScholarGoogle Scholar | 28424727PubMed |
Singh BK, Millard P, Whiteley AS, Murrell JC (2004) Unravelling rhizosphere-microbial interactions: opportunities and limitations. Trends in Microbiology 12, 386–393.
| Unravelling rhizosphere-microbial interactions: opportunities and limitations.Crossref | GoogleScholarGoogle Scholar | 15276615PubMed |
Stürmer SL, Kemmelmeier K, Moreira BC, Kasuya MCM, Pereira GMD, da Silva K (2018) Arbuscular mycorrhizal fungi (Glomeromycota) communities in tropical savannas of Roraima, Brazil. Mycological Progress 17, 1149–1159.
| Arbuscular mycorrhizal fungi (Glomeromycota) communities in tropical savannas of Roraima, Brazil.Crossref | GoogleScholarGoogle Scholar |
Tian B, Li J, Oakley TR, Todd TC, Trick HN (2016) Host-derived artificial microRNA as an alternative method to improve soybean resistance to soybean cyst nematode. Genes 7, 122
| Host-derived artificial microRNA as an alternative method to improve soybean resistance to soybean cyst nematode.Crossref | GoogleScholarGoogle Scholar |
Ward NL, Challacombe JF, Janssen PH, et al. (2009) Three genomes from the phylum Acidobacteria provide insight into the lifestyles of these microorganisms in soils. Applied and Environmental Microbiology 75, 2046–2056.
| Three genomes from the phylum Acidobacteria provide insight into the lifestyles of these microorganisms in soils.Crossref | GoogleScholarGoogle Scholar | 19201974PubMed |
Wu J, Yu M, Xu J, Du J, Ji F, Dong F, Li X, Shi J (2014) Impact of transgenic wheat with wheat yellow mosaic virus resistance on microbial community diversity and enzyme activity in rhizosphere soil. PLoS ONE 9, e98394
| Impact of transgenic wheat with wheat yellow mosaic virus resistance on microbial community diversity and enzyme activity in rhizosphere soil.Crossref | GoogleScholarGoogle Scholar | 24897124PubMed |
Zhang T, Jin Y, Zhao JH, Gao F, Zhou BJ, Fang YY, Guo HS (2016) Host-induced gene silencing of the target gene in fungal cells confers effective resistance to the cotton wilt disease pathogen Verticillium dahliae. Molecular Plant 9, 939–942.
| Host-induced gene silencing of the target gene in fungal cells confers effective resistance to the cotton wilt disease pathogen Verticillium dahliae.Crossref | GoogleScholarGoogle Scholar | 26925819PubMed |