High throughput phenotyping of root growth dynamics, lateral root formation, root architecture and root hair development enabled by PlaRoM
Nima Yazdanbakhsh A B and Joachim Fisahn AA Max Planck Institute of Molecular Plant Physiology, 14476 Potsdam Golm, Germany.
B Corresponding author. Email: yazdanbakhsh@mpimp-golm.mpg.de
This paper originates from a presentation at the 1st International Plant Phenomics Symposium, Canberra, Australia, April 2009.
Functional Plant Biology 36(11) 938-946 https://doi.org/10.1071/FP09167
Submitted: 2 July 2009 Accepted: 15 July 2009 Published: 5 November 2009
Abstract
Plant organ phenotyping by non-invasive video imaging techniques provides a powerful tool to assess physiological traits and biomass production. We describe here a range of applications of a recently developed plant root monitoring platform (PlaRoM). PlaRoM consists of an imaging platform and a root extension profiling software application. This platform has been developed for multi parallel recordings of root growth phenotypes of up to 50 individual seedlings over several days, with high spatial and temporal resolution. PlaRoM can investigate root extension profiles of different genotypes in various growth conditions (e.g. light protocol, temperature, growth media). In particular, we present primary root growth kinetics that was collected over several days. Furthermore, addition of 0.01% sucrose to the growth medium provided sufficient carbohydrates to maintain reduced growth rates in extended nights. Further analysis of records obtained from the imaging platform revealed that lateral root development exhibits similar growth kinetics to the primary root, but that root hairs develop in a faster rate. The compatibility of PlaRoM with currently accessible software packages for studying root architecture will be discussed. We are aiming for a global application of our collected root images to analytical tools provided in remote locations.
Additional keywords: Arabidopsis, growth profiling, video imaging.
Introduction
High throughput quantification of visible plant phenotypes has emerged as a valuable tool to characterise gene function, improvements of plant growth performance and biomass production. Therefore, an arsenal of tools that provide characterisation of diverse plant growth parameters has been developed. Initial approaches in the study of growth patterns in plants involved the use of rulers or digital callipers to determine the displacement of marked sectors. More recently, automated technologies using digital image processing have evolved to monitor organ development in a non-invasive manner. Most of these tools are appropriate to address very specific questions related to the complexity of whole-plant, organ or segmental growth. Because of this specification, only a small proportion of the dynamic and architectural parameters have been extracted from the sampled images. Thus, to optimise the information gain derived from kinematics of growing plants, it is desirable to develop a global network that links the accessibility to all presently available analytical tools.
Video imaging analysis has been used to determine the growth profiles of elongating plant organs such as pollen tubes, internodes, hypocotyls and leaves. However, less information is available about the growth responses of roots, mainly because of their inaccessibility. However, detailed growth analysis of roots has several advantages over other organs: (1) that growth is essentially unidirectional and strongly dependent on carbohydrates exported from the source leaves, and (2) that the local changes in water potential are likely to be less marked than in leaves. One approach to quantify root growth patterns uses calculations of the displacement of the root tip over time. This is achieved by capturing a series of time lapse records or marking root tip position on a transparent surface. To quantify root growth kinetics commercial software such as WINRHIZO (Arsenault et al. 1995), OPTIMAS analysis software (Media Cybernetics, Bethesda, MD, USA) or IMAGE J (Abramoff et al. 2004) was introduced to assess defined growth parameters. Several laboratories are even providing freely available software packages to analyse root architecture and growth (Basu et al. 2007; Armengaud et al. 2009).
Previous approaches have shown that there are rapid adaptations of growth in responses to modulation in light intensities and periods (Aguirrezabal et al. 1994; Muller et al. 1998; Nagel et al. 2006). Furthermore, they demonstrated that there are different growth zones at the root tip, and that these are differentially affected by different treatments (Walter et al. 2003). Recent studies have quantified differences in root architecture (Armengaud et al. 2009). By combining curvature measurements with root diameter, the differential growth ratio between the greater and lesser curvature edges of a bending root was calculated by KineRoot (Basu et al. 2007). However, information on how roots respond to combined stimuli; varying carbohydrate status, different ratios of nutrients, nitrogen, phosphate or sulfur and combined nutritional and abiotic stress is sparse. Early studies by time lapse recordings with cherry (Prunus avium) root tips demonstrated significant increase in the averaged rate of growth in the night compared with the day (Head 1965). The root elongation rate of rice (Oryza sativa L.) seemed to show a rhythmic pattern, but the timing of the maxima and minima was not robust (Iijima et al. 1998). More recent studies with rice and tobacco found similar root tip velocities in the day and night in a 12-h photoperiod (Walter et al. 2002; Walter and Schurr 2005). However, the root elongation kinetics of the model plant Arabidopsis thaliana (L.) Heynh is not recorded in published literature.
The most recent platforms for non-invasive analysis of root growth provide accurate and reliable results with high temporal or spatial resolution, but they are time-consuming, often targeted and have a low throughput. However, the effect of environmental stimuli on biological processes especially growth and development is well known. Hence, it is difficult to reveal reliable patterns and detect small changes of root elongation from currently available data. We here describe a range of applications of a recently developed plant root monitoring platform (PlaRoM) that enables high throughput monitoring of growing seedlings with high spatial and temporal resolution and detection of root elongation profiles.
Materials and methods
Plant material and cultivation procedure
Seeds of Arabidopsis thaliana (L.) Heynh wild-type accession (Col0) were surface-sterilised for 20 min with 10% sodium hypochlorite solution containing 0.1% surfactant (Triton X-100). After sterilisation, seeds were rinsed several times with sterile water and plated on the surface of solid nutrient agar (7.0% m/v) supplemented with half-strength MS medium (Murashige and Skoog 1962) (M02 555, pH 5.6; Duchefa, Haarlem, The Netherlands). After 4 days stratification, Petri dishes were placed vertically in the phytotron (21°C constant day and night temperature, 100 µmol m–2 s–1 photon flux density). Photoperiod was set to 12/12 h day/night. At day 9, seedlings that had developed roots of at least 1 cm were selected and distributed equally in a row 3 cm down the top on the surface of solid medium filled in 120 × 120 mm rectangular Petri dishes. For the extended night experiment two growth media were used for different Petri dishes, one of which was complemented by 0.01% sucrose. Sample plates were kept in the phytotron for 2 more days.
Results and discussion
Plant root monitoring platform (PlaRoM)
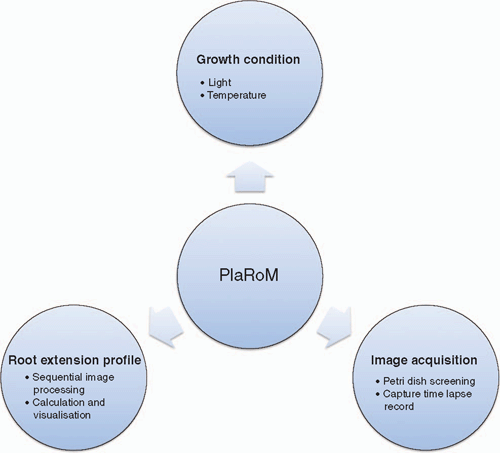
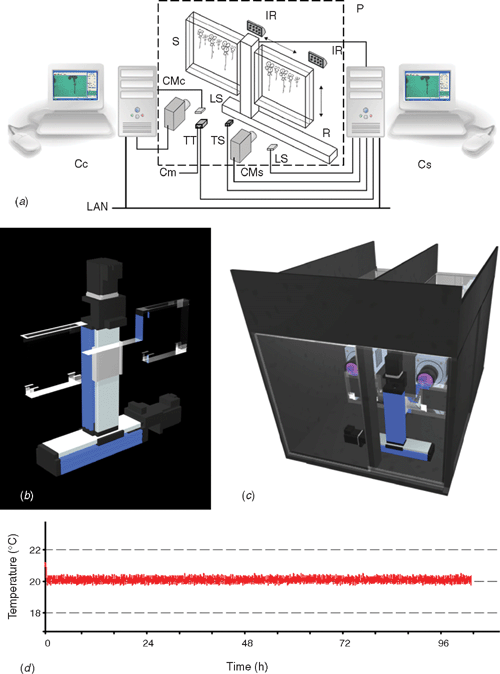
Plant phenotyping in a non-invasive way requires video imaging where the quality of images defines the resolution of the measurement. High throughput characterisation of growth, however, requires robotised equipment for imaging and adequate software application for processing these records. However, due to modulation of plant growth in response to abiotic stimuli (Franklin 2008; Walter et al. 2009), environmental conditions must be controlled and recorded in any growth study. To fulfil these requirements, we have developed a plant root monitoring platform (PlaRoM) that enables high resolution monitoring of up to 50. A. thaliana seedlings growing in controlled growth condition. PlaRoM controls the growth condition in which sample seedlings are growing, acquires images of growing seedlings and finally calculates the root extension profiles and plots the results over time (Fig. 1). Hence, the imaging platform consists of a custom designed phytochamber and the enclosed measuring head (Fig. 2). Two software applications were developed for PlaRoM. While the imaging application controls the imaging platform (constant growth condition and image acquisition), the root extension profiling application calculates the growth profiles from obtained images and plots the results.
|
|
Controlled constant growth condition
A custom designed phytochamber housed the measuring head of PlaRoM (Fig. 2c). The top and front layers of this chamber were made up of transparent material. Hence, the photosynthetic, as well as imaging light sources, were placed outside the chamber to reduce heat production therein. The light intensity in the chamber at the surface of the leaves of the seedlings was typically 70–90 µmol m–2 s–1. Control over temperature regimes was achieved by combination of a USB temperature sensor and a digital thermometer and thermostat regulating a cooling device. Further, operation of three fans circulated the air inside the phytochamber and, thus, prevented the development of a temperature gradient. A module in the imaging software application set the temperature through the digital thermometer and thermostat with 0.5°C accuracy (Fig. 2d). In addition, digital light sensors enabled the recording of photon flux densities at the start of each screening of the Petri dishes.
Image acquisition
In order to study dynamics of root growth, high resolution time lapse records of several individuals with relative short time intervals are needed. Hence, the PlaRoM imaging platform screened the surface of two sample plates and captured time lapse records of the seedlings growing therein. In detail, a custom designed sample stage housed two Petri dishes, each monitored by a camera-microscope unit. The imaging light was provided by infrared light sources, each placed in front of a camera-microscope unit (Fig. 2a). The sample stage was mounted on a screening robot which provided its motion (Fig. 2b). The PlaRoM imaging software application controlled the screening of the Petri dish and capturing of time lapse records (a brief description of the robotised platform for monitoring a single Petri dish is outlined by Yazdanbakhsh and Fisahn 2007a).
Petri dish screening
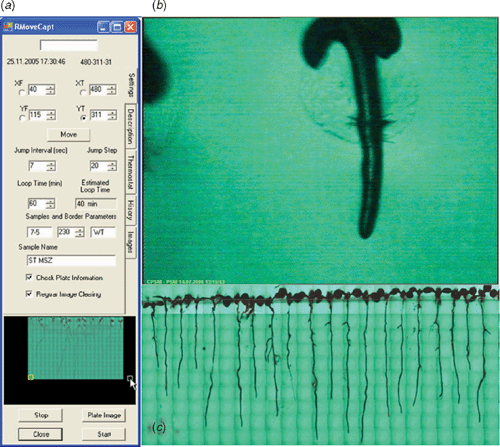
Central to the multi parallel recording of many roots was the screening robot. This robot consisted of two perpendicularly arranged linear stages (Fig. 2a, b). Stepwise movement of the robot arm, controlled by the imaging software application, enabled the movement of the sample stage in a plane. This automated motion enabled screening of a user defined area (corresponding to the rectangle [(XF, YF), (XT, YT)] in robot arm coordinate system, Fig. 3a) over the surface of each Petri dish over several days.
|
Capturing time lapse records
EasyGrab, a software package that operates the frame grabbing card, provides a 768 × 576 pixel live video stream from the cameras. With the presently used magnification set by the microscopes, the video stream covers a 4.58 × 3.33 mm area over the surface of the Petri dish. Current magnification enables capturing images with 5.96 × 5.78 µm pixel–1 resolution (Fig. 3b). However, higher spatial resolution is possible by greater zoom. Stepwise movements of the robot arm were applied at user defined time intervals of at least 5 s (Fig. 3a). To assure the highest degree of reliability in detection of the root tip positions, reduce the effect of noise (mainly optical noise in the background), and obtain smooth curves, three time lapse records were captured at each position with 800-ms intervals. These records are stored in a subfolder of the measurement folder, labelled by the position of the viewing frame in robot arm coordinate system, and are used for calculation of root extension profiles. To facilitate the root tip detection process each time lapse record is stored under a special filename format including its capturing time.
While capturing time lapse records, a module in imaging software application built an image of the whole screened area, and this was stored at the end of each completed screen. In detail, upon capture, a 200 × 200 pixel ‘scaled-down’ copy of one of the obtained time lapse records was placed in position of a composed image relative to the corresponding view frame coordinates (Fig. 3c). These images present the growth status of all individuals during the measurement period.
Root extension profiling
The root extension profiling software application analysed the time lapse records and provided the growth velocity profiles. Modules embedded in this application were developed for sequential image processing, calculation of the root extension patterns and visualisation of the obtained growth profiles.
Sequential image processing
The sequential image processing module applied the root tip detection algorithm (Yazdanbakhsh and Fisahn 2007b) to a stack of images captured from each individual root. In detail, the user defined the root tip detection parameters and prepared a list of the indices of images showing the root tip in each view frame. For a measurement applied over 4 days with 30 min screening periods, the image stack contained ~600 records. This module applied the root tip detection algorithm to all corresponding records and detected the root tip position. Finally, the coordinates of the detected root tip together with the times of dawn and dusk were stored in an XML library under a name assigned to that individual.
Visualisation of root extension profiles
This module calculated the extension profile of an individual root, and plotted the values over time. Extension profiles were obtained from displacements of each individual root tip in consecutive time points during the measurement. This application provided plots of the calculated root extension profiles.
Investigation of root extension profiles by PlaRoM
PlaRoM provided a powerful tool to investigate the dynamics of root extension profiles and marked higher throughput, spatial and temporal resolution over presently available methods to quantify root growth dynamics. High throughput monitoring of up to 50 individuals is performed simultaneously owing to the applied robotised motion system. Most of the previously described devices focus on the detection of one individual root. Screening two Petri dishes in 30 min time periods provides high resolution root extension profiles. Furthermore, the constant controlled growth conditions enable investigation of root extension patterns in different environmental conditions. Subsequently, we will demonstrate several applications of PlaRoM.
Dynamics of primary root extension
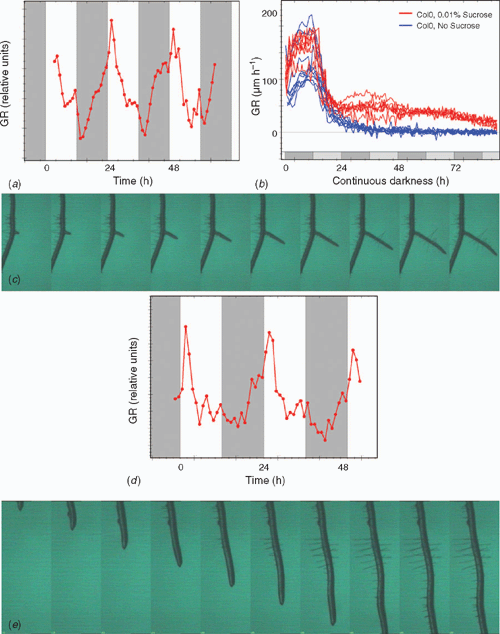
The high spatial and temporal resolution provided by the platform enables analysis of root extension rate over short time intervals. Furthermore, the platform can be used to study dynamics of root elongation in different light regimes. Root extension kinetics of A. thaliana primary roots (Fig. 4a) demonstrated the existence of similar pattern compared with that of previously described leaf growth profiles (Wiese et al. 2007). Root elongation rate declined during illumination, while a recovery emerged towards the end of this period. The main part of the night was characterised by an increase in root growth activity. We have intensively studied extension kinetics of A. thaliana primary roots in different light protocols which will be published soon (Smith and Stitt 2007; N. Yazdanbakhsh and J. Fisahn, unpubl. data).
|
Root extension patterns in different growth media
PlaRoM enables the study of root elongation in different growth media, especially with its option to screen two Petri dishes simultaneously. Figure 4b displays the relaxation kinetics of A. thaliana wild-type seedlings during 90 h of darkness and its modulation due to 0.01% extracellular sucrose (blue and red traces, n = 11, n = 7, respectively). Inhibition of root elongation in Col0 seedlings started shortly after the expected dawn. In the absence of sucrose, continuation of this gradual decrease led to complete cessation of growth after ~24 h of extension of the night. In the presence of 0.01% sucrose, however, 8–10 h after extension of the night slight recovery of root extension was detected (Fig. 4b). We note that root elongation increased in the anticipated night, and gained a local maximum at the anticipated dawn. This finding indicates that in continues darkness the presence of 0.01% sucrose modifies the relaxation kinetics of Arabidopsis roots in a characteristic way. This measurement demonstrates the ability of PlaRoM to detect the modulation of root elongation in response to very small amounts of extracellular carbohydrate.
Comparison of root extension profiles among different genotypes
PlaRoM can be used to study root extension profiles of several genotypes, including mutants. Hence, it is a suitable tool to investigate the role of certain genes (or the related physiological pathways) in the growth process. In particular, we have already investigated the defected root elongation patterns of a set of starch mutants, as well as circadian clock mutants in different light protocols including free running conditions. Among starch mutants pgm, sex1, mex1, dpe1 and dpe2 were investigated, all of which present different levels of nocturnal growth inhibition. The strongest defect of up to 80% inhibition of growth at the end of the night (Smith and Stitt 2007) is exhibited by pgm, while mex1 and dpe1 exhibit quite similar growth pattern compared with the wild type (data not shown). Furthermore, root extension patterns measured from circadian clock mutants (elf3, ef4 and the cca1-lhy double mutant) in different light protocols including free running conditions display deviations from that of the wild type. These deviations were detected in the second part of the day and night period. All mutants display inhibition of growth towards dawn and an increase of growth activity towards dusk. Furthermore, in free running experiment of continuous light, they present different time constants compared with that of the wild type, and elf3 mutant displays arrhythmic root extension pattern. Detailed investigation of the root extension patterns of Arabidopsis circadian clock mutants and their modification due to extracellular sucrose will be published separately.
Kinetics of lateral root development
Lateral root (LR) formation is of great agronomic significance, since highly branched LRs could greatly increase the absorptive area of the root system that uptakes water and nutrients and provides physical support to the plant. Thus, the development of an optimal LR system is a key factor for the vigour of the plant in various conditions (Malamy and Benfey 1997). Root system architecture is a highly plastic trait, and varies both within and among species (Osmont et al. 2007). However, the basic root system morphology and its plasticity are controlled by inherent genetic factors. These mediate the modification of root system architecture, mostly at the level of root branching, in response to a suite of biotic and abiotic factors (Osmont et al. 2007). The PlaRoM imaging platform screens each sample plate for several days. Hence, any developmental process within each Petri dish can be followed from its initial stage. A series of images displaying the development of a lateral root is presented in Fig. 4c. LR development occurs at locations distal to the root tip, and progresses in parallel to elongation of the main root. Extension kinetics of lateral roots can be calculated from the displacement of the lateral root tip (Fig. 4d). Similar to the primary root, maximal growth activity was detected in the morning which decreases during the light period and increases during the major part of the night.
Root hair development
Root hairs are tubular structures that arise from root epidermal cells. The functional absorbing surface of a root hair is at the tip only. Hence, root hairs function to increase the apparent diameter of the root (Segal et al. 2008). A low-pH (pH 4) medium induced root hair formation in lettuce (Lactuca sativa L.) seedlings (Dolan and Davies 2004). Results suggest that factors essential to the formation of root hairs may be present in the shoot. The PlaRoM imaging platform provides high resolution images of root hair formation as well. Figure 4e presents a typical image sequence showing the development of root hairs. In this example, root hair development requires 12–15 h. This is a much faster time scale than the lateral roots. Root hair initiation has been reported to be controlled by the same genes that induce trichome formation (Schellmann et al. 2007; Ishida et al. 2008). Therefore, future application of the platform to mutants in trichome formation will provide novel phenotypes in root hair development.
Compatibility and application of PlaRoM images to public image processors
The image files presenting the screened area over the surface of the Petri dish can be used by currently available software applications for investigation of root architecture. Depending on the capability of these programs in detection of big bitmap files, the image files with higher pixel resolution can also be produced by the imaging platform. In detail, higher magnification through the microscope enables detailed study of root hair development. Figure 5 presents the application of EZ-Rhizo (Armengaud et al. 2009) to one of the images collected by the PlaRoM showing the growth status of Col0 and two cell wall mutants with proteins of unknown function growing in a Petri dish (n = 11, n = 6, n = 6, respectively). The software application detects the roots and provides the length and angle of main and lateral roots, as well as total number of lateral roots (Fig. 5; Armengaud et al. 2009).
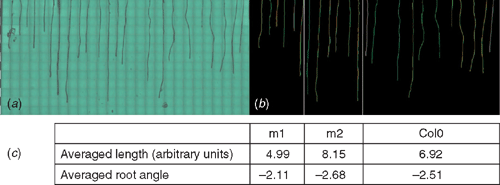
|
Outlook
Plant phenomics is a rapidly emerging area of research that combines the development of new imaging technologies with plant physiological and genomics data. Although electrical and mechanical engineering has advanced in many industrial branches, only limited progress has been reported on automated plant phenotyping and integration of these results into the framework of systems biology. Therefore, a new generation of scientific work is needed that incorporate expertise in systems analysis, electrical engineering and plant physiology. Development of PlaRoM provides one step into this next generation. The imaging platform enables simultaneous monitoring of several root tips over long time, and provides a great advantage over the currently available root monitoring systems. PlaRoM reveals the root extension pattern of different genotypes in constant growth conditions. Furthermore, it enables to study the modulation of these patterns due to different light protocols, growth media, temperature regimes as well as presence of different bacteria in growth medium. Investigation of root extension patterns of more circadian clock related genes and their modulation due to extracellular sucrose will be published soon. In addition, effect of short light pulses, as well as different temperature regimes on root extension profiles of Arabidopsis will be studied in near future. With the possibility of applying already available software applications to the images obtained from PlaRoM imaging platform, we would like to study root architectures and development of lateral roots with high spatial and temporal resolution. Furthermore, obtained images enable us to accomplish a detailed study of root hair formation.
To optimise the availability and, thus, use of already existing data mining algorithms, we suggest a global network of digital image processing in the field of plant research. This should include a joint video image database of developing and growing plant organs to facilitate detailed mining of currently available digital imaging data and link those to genomics, proteome an metabolite databases.
Acknowledgements
This work was supported by the Max Planck Society and by a contract to N. Y. We thank Professor Dr M Stitt (MPI Potsdam, Germany) for valuable discussion during the development of PlaRoM.
Abramoff MD,
Magelhaes PJ, Ram SJ
(2004) Image processing with ImageJ. Biophotonics International 11, 36–42.

Aguirrezabal LAN,
Deleens E, Tardieu F
(1994) Root elongation rate is accounted for by intercepted PPFD and source–sink relations in-field and laboratory-grown sunflower. Plant, Cell & Environment 17, 443–450.
| Crossref | GoogleScholarGoogle Scholar |

Armengaud P,
Zambaux K,
Hills A,
Sulpice R,
Pattison RJ,
Blatt MR, Amtmann A
(2009) EZ-Rhizo: integrated software for the fast and accurate measurement of root system architecture. The Plant Journal 57, 945–956.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Arsenault JL,
Pouleur S,
Messier C, Guay R
(1995) Win-RHIZO, a root-measuring system with a unique overlap correction method. HortScience 30, 906.

Basu P,
Pal A,
Lynch JP, Brown KM
(2007) A novel image-analysis technique for kinematic study of growth and curvature. Plant Physiology 145, 305–316.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Dolan L, Davies J
(2004) Cell expansion in roots. Current Opinion in Plant Biology 7, 33–39.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Franklin KA
(2008) Light and temperature signal crosstalk in plant development. Current Opinion in Plant Biology 12, 1–6.
| PubMed |

Head GC
(1965) Studies of diurnal changes in cherry root growth and nutational movements of apple root tips by time-lapse cinematography. Annals of Botany 29, 219–224.

Iijima M,
Oribe Y,
Horibe Y, Kono Y
(1998) Time lapse analysis of root elongation rates of rice and sorghum during the day and night. Annals of Botany 81, 603–607.
| Crossref | GoogleScholarGoogle Scholar |

Ishida T,
Kurata T,
Okada K, Wada T
(2008) A genetic regulatory network in the development of trichomes and root hairs. Annual Review of Plant Biology 59, 365–386.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Malamy JE, Benfey PN
(1997) Down and out in Arabidopsis: the formation of lateral roots. Trends in Plant Science 2, 390–396.
| Crossref | GoogleScholarGoogle Scholar |

Muller B,
Stosser M, Tardieu F
(1998) Spatial distributions of tissue expansion and cell division rates are related to irradiance and to sugar content in the growing zone of maize roots. Plant, Cell & Environment 21, 149–158.
| Crossref | GoogleScholarGoogle Scholar |
CAS |

Murashige T, Skoog F
(1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiologia Plantarum 15, 473–497.
| Crossref | GoogleScholarGoogle Scholar |
CAS |

Nagel KA,
Schurr U, Walter A
(2006) Dynamics of root growth stimulation in Nicotiana tabacum in increasing light intensity. Plant, Cell & Environment 29, 1936–1945.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Osmont KS,
Sibout R, Hardtke CS
(2007) Hidden branches: developments in root system architecture. Annual Review of Plant Biology 58, 93–113.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Schellmann S,
Hulskamp M, Uhrig J
(2007) Epidermal pattern formation in the root and shoot of Arabidopsis. Biochemical Society Transactions 35, 146–148.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Segal E,
Kushnir T,
Mualem Y, Shani U
(2008) Water uptake and hydraulics of the root hair rhizosphere. Vadose Zone Journal 7, 1027–1034.
| Crossref | GoogleScholarGoogle Scholar |

Smith AM, Stitt M
(2007) Coordination of carbon supply and plant growth. Plant, Cell & Environment 30, 1126–1149.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Walter A,
Spies H,
Terjung S,
Kusters R,
Kirchgessner N, Schurr U
(2002) Spatio-temporal dynamics of expansion growth in roots: automatic quantification of diurnal course and temperature response by digital image sequence processing. Journal of Experimental Botany 53, 689–698.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Walter A,
Feil R, Schurr U
(2003) Expansion dynamics, metabolite composition and substance transfer of the primary root growth zone of Zea mays L. grown in different external nutrient availabilities. Plant, Cell & Environment 26, 1451–1466.
| Crossref | GoogleScholarGoogle Scholar |
CAS |

Walter A, Schurr U
(2005) Dynamics of leaf and root growth: endogenous control versus environmental impact. Annals of Botany 95, 891–900.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Walter A,
Silk WK, Schurr U
(2009) Environmental effects on spatial and temporal patterns of leaf and root growth. Annual Review of Plant Biology 60, 279–304.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Wiese A,
Christ MM,
Virnich O,
Schurr U, Walter A
(2007) Spatio-temporal leaf growth patterns of Arabidopsis thaliana and evidence for sugar control of the diel leaf growth cycle. New Phytologist 174, 752–761.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |



