The structure and activity of nodulation-suppressing CLE peptide hormones of legumes
April H. Hastwell A , Peter M. Gresshoff A and Brett J. Ferguson A BA Centre for Integrative Legume Research, School of Agricultural and Food Sciences, The University of Queensland, St Lucia, Brisbane, Qld 4072, Australia.
B Corresponding author. Email: b.ferguson1@uq.edu.au
Functional Plant Biology 42(3) 229-238 https://doi.org/10.1071/FP14222
Submitted: 8 August 2014 Accepted: 3 November 2014 Published: 12 December 2014
Abstract
Legumes form a highly-regulated symbiotic relationship with specific soil bacteria known as rhizobia. This interaction results in the de novo formation of root organs called nodules, in which the rhizobia fix atmospheric di-nitrogen (N2) for the plant. Molecular mechanisms that regulate the nodulation process include the systemic ‘autoregulation of nodulation’ and the local nitrogen-regulation of nodulation pathways. Both pathways are mediated by novel peptide hormones called CLAVATA/ESR-related (CLE) peptides that act to suppress nodulation via negative feedback loops. The mature peptides are 12–13 amino acids in length and are post-translationally modified from the C-terminus of tripartite-domain prepropeptides. Structural redundancy between the prepropeptides exists; however, variations in external stimuli, timing of expression, tissue specificity and presence or absence of key functional domains enables them to act in a specific manner. To date, nodulation-regulating CLE peptides have been identified in Glycine max (L.) Merr., Medicago truncatula Gaertn., Lotus japonicus (Regel) K.Larsen and Phaseolus vulgaris L. One of the L. japonicus peptides, called LjCLE-RS2, has been structurally characterised and found to be an arabinosylated glycopeptide. All of the known nodulation CLE peptides act via an orthologous leucine rich repeat (LRR) receptor kinase. Perception of the peptide results in the production of a novel, unidentified inhibitor signal that acts to suppress further nodulation events. Here, we contrast and compare the various nodulation-suppressing CLE peptides of legumes.
Additional keywords: autoregulation of nodulation, legume nodulation, nitrate-regulation of nodulation, nodule, plant peptide signalling, symbiosis.
Introduction
The common agricultural practice of using nitrogen-based fertilisers to increase crop yields has been highly successful in generating sufficient food for the world’s ever-growing population. It has been a major part of the ‘green revolution’ instigated more than 50 years ago. However, adverse economical and ecological consequences are beginning to outweigh the benefits of nitrogen fertiliser use (Erisman et al. 2008; Sutton et al. 2011; Jensen et al. 2012).
Symbiotic nitrogen fixation represents an alternative to chemical nitrogen fertiliser use. It involves a relationship mainly formed between plant species of the family Fabaceae, commonly known as legumes, and soil bacteria, collectively referred to as rhizobia. Major legume crop and pasture species include soybean, pea, common bean, clover, cowpea, medic, chickpea, lentil and peanut. Biological nitrogen fixation from this legume–rhizobia relationship currently results in ~50–70 Tg of nitrogen added into global agricultural systems each year (Herridge et al. 2008; Jensen et al. 2012).
The legume-rhizobia relationship is signified by the formation of a new plant organ, called the nodule. Nodule development is orchestrated by a complex signalling interaction (Ferguson and Mathesius 2003, 2014; Ferguson et al. 2010; Desbrosses and Stougaard 2011; Oldroyd 2013). Once formed, the nodule acts to house the rhizobia that provide the plant with a useable form of reduced nitrogen (namely ammonia) using a specialised enzyme complex to ‘fix’ un-reactive atmospheric di-nitrogen gas (N2). In return, the rhizobia are provided with a carbon source derived from photosynthesis, predominately malate (Udvardi et al. 1988). In addition to increasing current crop yields, this process is exploited in agriculture to improve the nitrogen content and structure of soils by using legumes as rotation crops (Jensen et al. 2012).
Control of legume nodule numbers
Nodulation is costly to the host plant in terms of resources; as a result, the plant has developed both local and systemic mechanisms to control its nodule numbers (Delves et al. 1986; Gresshoff and Delves 1986; recently reviewed by Reid et al. 2011b). Local control mechanisms responding to high soil nitrate directly prevent or delay nodule development (Carroll et al. 1985a; Reid et al. 2011a). A systemic control mechanism, called the ‘autoregulation of nodulation’ (AON), is closely associated with the nitrate regulatory pathway, but is induced by rhizobia, not nitrate, and acts systemically through the shoot, rather than locally in the root (Kosslak and Bohlool 1984; Delves et al. 1986; Gresshoff and Delves 1986; Reid et al. 2011a, 2011b).
The AON process begins with the production of a root-derived signal (Gresshoff and Delves 1986), which is expressed in response to a transcription factor, called NIN, involved in cortical cell division during early nodulation events (Soyano et al. 2014). This signal, formerly called ‘Q’ (Gresshoff and Delves 1986), is now known to be a CLAVATA/Embryo surrounding region (ESR) related (CLE) peptide. To date, CLE peptide-encoding genes having a role in nodulation have been identified in Glycine max (L.) Merr. (soybean), Medicago truncatula Gaertn., Lotus japonicus (Regel) K.Larsen and Phaseolus vulgaris L. (common bean) (Fig. 1; Okamoto et al. 2009, 2013; Mortier et al. 2010, 2012; Lim et al. 2011; Saur et al. 2011; Reid et al. 2011a, 2013; Ferguson et al. 2014). Recent biochemical advances have enabled the isolation and identification of one of the nodulation CLE peptides of L. japonicus, called LjCLE-RS2. The mature signal of this CLE peptide is 13 amino acids in length, is derived from a much larger prepropeptide and is post-translationally modified with three β 1-2 linked arabinose moieties at Hyp7 (Okamoto et al. 2013).

|
The nodulation-suppressing CLE peptide signal is exported from the root and transported via the xylem by an unknown mechanism (Okamoto et al. 2013) to the leaf phloem parenchyma (Nontachaiyapoom et al. 2007) where it is perceived by a leucine-rich repeat serine-threonine receptor kinase (LRR RK), called GmNARK in soybean, LjHAR1 in L. japonicus, MtSUNN in M. truncatula, PsSYM29 in Pisum sativum L. (pea), GsNARK in Glycine soja Siebold & Zucc., and PvNARK in common bean (Krusell et al. 2002; Nishimura et al. 2002; Searle et al. 2003; Schnabel et al. 2005; Ferguson et al. 2014). These LRR RKs may act in a complex with other receptors to perceive the CLE peptide ligand. This includes factors such as LjCLAVATA2/PsCLAVATA2 and LjKLAVIER (Miyazawa et al. 2010; Krusell et al. 2011). Additional research has identified other factors that may interact with the LRR RK directly, or function downstream of it, to relay the perception of the signal and trigger downstream signalling events. This includes the kinase-associated protein phosphatases, GmKAPP1 and GmKAPP2 (Miyahara et al. 2008), the putative ubiquitin fusion degradation protein, GmUFD1a (Reid et al. 2012) and the root-acting F-box protein, TOO MUCH LOVE, LjTML (Magori et al. 2009; Takahara et al. 2013). Following the perception of the nodulation CLE peptide signal, a shoot-derived inhibitor (SDI) signal is produced and transported to the roots, likely via the phloem, to inhibit further nodulation development (Delves et al. 1986; Lin et al. 2010, 2011; Reid et al. 2011b; Sasaki et al. 2014). Recent studies in L. japonicus have indicated a role for cytokinin as a potential SDI-candidate in AON (Sasaki et al. 2014).
The gene encoding for the LRR RK is expressed in both shoot and root tissues (Krusell et al. 2002; Nishimura et al. 2002; Searle et al. 2003; Schnabel et al. 2005; Nontachaiyapoom et al. 2007) and plants having mutations in it exhibit both supernodulation (due to a lack of AON control) and nitrate-tolerant nodulation phenotypes (e.g. Carroll et al. 1985a, 1985b). Grafting studies using soybean have demonstrated that the GmNARK LRR RK is required for both AON in the shoot (Delves et al. 1986; Reid et al. 2011a) and nitrate-regulation of nodulation in the root (Reid et al. 2011a). Similar to AON, the nitrate regulation of nodulation mechanism in soybean begins with the production of a CLE peptide that is predicted to be perceived by GmNARK. However, unlike AON, this CLE peptide, called GmNIC1, responds to nitrate, not rhizobia, and acts locally in the root, not systemically in the shoot. These findings helped to confirm that there are two independent pathways controlling nodulation: the systemic rhizobia-induced AON pathway and the local nitrate-induced regulation of nodulation pathway (reviewed by Reid et al. 2011b). No candidates for the root-derived inhibitor (RDI) have been identified to date, but as it is produced downstream of GmNARK, it may be similar to, or even the same as, the SDI signal in AON.
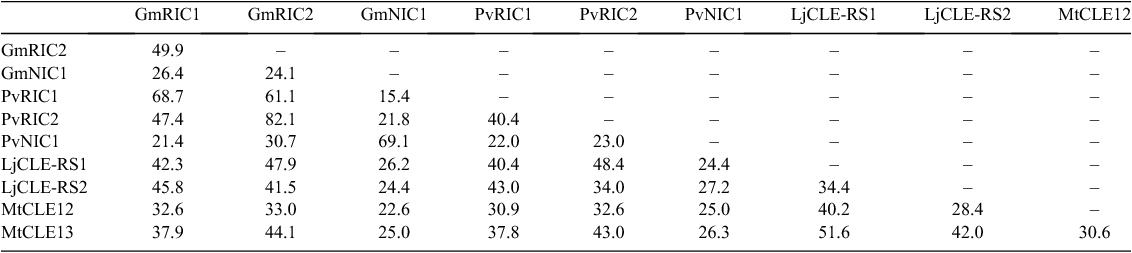
Unlike soybean, L. japonicus and M. truncatula appear to have overlapping local and systemic molecular mechanisms that act to regulate nodulation in response to both rhizobia and nitrate (Okamoto et al. 2009; Mortier et al. 2010). Although the reason for this difference amongst species is unknown, it is likely that it relates to genomic duplication events undergone in soybean that have enabled genetic divergence and the development of new molecular signals and mechanisms through the process of neofunctionalisation (Schmutz et al. 2010). This may also explain why soybean has three functional CLE peptides that are known to regulate nodule numbers, in addition to three homeologous (duplicate) copies that may have no-, reduced- or diverged-function (Reid et al. 2011a), whereas L. japonicus and M. truncatula appear to have only two such peptides (Table 1). Interestingly common bean, which shared a duplication event with soybean 53 million years ago, has orthologous copies of the three soybean CLE peptide genes, but lacks the duplicate copies of each of these genes as a result of not undergoing the more recent genome duplication event approximately 13 million years ago (Ferguson et al. 2014).

|
The CLE peptides that act as a trigger for AON and nitrate-regulation of nodulation belong to a large group of heavily processed, cysteine-poor secreted plant peptides related to AtCLV3 in Arabidopsis thaliana (L. Heynh.) (Matsubayashi 2014). AtCLV3 functions in the CLAVATA pathway to regulate the shoot apical meristem stem cell population. It acts as a ligand to a receptor-complex involving AtCLV1, AtCLV2 and AtCORYNE (Ogawa et al. 2008). The AtCLV1 receptor is a LRR RK that is highly similar in structure to the LRR RKs that are central to nodulation control. Other Arabidopsis CLE peptides of note that are similar to the nodulation CLE peptides of legumes include AtCLE1 to AtCLE7, which have roles in root architecture and development (Cock and McCormick 2001; Strabala et al. 2006; Oelkers et al. 2008; Araya et al. 2014).
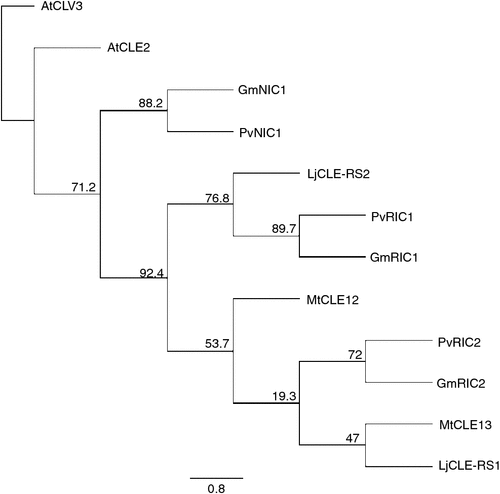
The mechanisms controlling nodulation in legumes are highly conserved, as demonstrated by the interspecific function of AON CLE peptides from soybean in common bean and from M. truncatula in pea (Osipova et al. 2012; Ferguson et al. 2014). There are, however, many differences in the sequences, structures and inducing factors of the various nodulation CLE peptides that allow for specificity of function (Fig. 1; Tables 1, 2). These similarities and differences, and how they impact on nodule suppression, are reviewed here.
|
Key functional domains of CLE peptides
Mature CLE peptide signals are derived from prepropeptides consisting of 3–4 domains: an N-terminal signal peptide, a variable region and a CLE domain, with some also having a C-terminal extension (Fig. 1). Sequence similarities amongst the nodulation CLE prepropeptides shows the orthologous copies are most similar (Fig. 2; Table 2); however, it is likely that similarities and differences in the individual domains are most critical for driving specificity. Here we discuss the function, conservation and importance of each domain, particularly in respect to their role in the suppression of nodulation.
|
Signal peptide
The N-terminal hydrophobic signal peptide (also referred to as a transit peptide) is widely thought to be responsible for exporting the prepropeptide out of the cell (la Cour et al. 2004; Lim et al. 2011). It is ~30 amino acids in length and is critical to the specificity of the peptide (Fletcher et al. 1999; Reid et al. 2013). This domain has a role in exporting the AtCLV3 propeptide into the extracellular space (Rojo et al. 2002). A similar role for this domain is predicted for the nodulation CLE peptides.
Amongst the known nodulation CLE prepropeptides, the signal peptide domain has a 37% pairwise identity and contains a leucine-rich motif (Fig. 1), commonly observed in exported proteins (la Cour et al. 2004). Also present within this domain is a conserved motif of five amino acids (TLQAR; Table 1), which is predicted to be a site of cleavage (Okamoto et al. 2009). It has been noted that GmNIC1, PvNIC1 and MtCLE12 show lower conservation of amino acid residues within this motif. They are also the only known nodulation CLE peptides to lack the C-terminal extension domain (Table 1; Fig. 1). Outside of this motif, conserved sequence residues within the signal peptide can be seen amongst predicted orthologues of the nodulation CLE peptides (Fig. 1; Reid et al. 2011a; Ferguson et al. 2014).
Variable region
The functional importance of the variable domain, the least conserved of the four domains, remains unknown (Ni and Clark 2006; Meng et al. 2010; Reid et al. 2013). Indeed, AtCLV3 shows function without this domain (Fiers et al. 2006). The size of the domain is also highly variable (31–50 amino acids). However, recognition and cleavage immediately before the Arg1 residue of the CLE domain requires at least four to five residues of the variable domain to be present for correct processing of AtCLV3 (Kondo et al. 2008; Ni et al. 2011; Xu et al. 2013). An additional amino acid at residue 39 within the variable domain of AtCLV3 is also predicted to be a cleavage site (Xu et al. 2013).
There are no residues that are 100% conserved across the variable domain between the known nodulation CLE peptides (Fig. 1), although it shows a 19.4% pairwise identity and, as with other domains, residues are conserved between orthologues. This is particularly evident between the nodulation CLE peptides of the closely-related bean and soybean species (Fig. 1; Ferguson et al. 2014).
CLE domain
The CLE domain, from which the peptide is named, denotes the mature/active peptide sequence. It is located at the C-terminus and is the most conserved region (Cock and McCormick 2001; Oelkers et al. 2008). The consensus amino acid sequence of the nodulation-suppressing CLE peptides is RL(A/S)PGGPDPQHN(X) (Fig. 1). The domain is 12 or 13 amino acids in length and contains 50% identical sites, with 77.4% (12 amino acids) and 75.2% (13 amino acids) pairwise identity between the known nodulation CLE peptides. LjCLE-RS2 of L. japonicus is the only structurally-confirmed nodulation CLE peptide, and is 13 amino acids in length (Okamoto et al. 2013). However, the nitrate-induced GmNIC1 peptide of soybean and its orthologue in bean, PvNIC1, have a stop codon at position 13 and therefore can only be 12 amino acids in length (Fig. 1; Reid et al. 2011a; Ferguson et al. 2014). This may influence their functional properties, such as their apparent lack of long distance transport.
Notably, GmRIC1, PvRIC1, GmRIC2, and PvRIC2 are the only nodulation CLE peptides known to contain an Ala residue at position 3 of the CLE domain, presumably a result of polyploidisation and subsequent species divergence amongst the legumes (Figs 1, 2; Stefanović et al. 2009; Schmutz et al. 2010). There are four other residues within the CLE domain of the known nodulation CLE peptides that contain sequence divergence from the consensus sequence: Gly5 > Glu5 (GmRIC1 and PvRIC1) or Ala5 (MtCLE13); Asp8 > Asn8 (MtCLE12); Pro9 > His9 (MtCLE12) or Gln9 (GmNIC1); and Gln10 > His10 (GmRIC1 and PvRIC1) or Ile10 (MtCLE12) (Fig. 1). It is not yet known how the activity of the CLE peptide is affected by these sequence divergences. Only the one at position 8 in MtCLE12 is predicted to be critical for function (i.e. the suppression of nodulation) based on site-directed mutagenesis work using soybean (Reid et al. 2013); however, this nonsynonymous substitution from an uncharged asparagine to a negatively charged aspartic acid is conservative and may not affect activity.
Recent research has indicated that, despite sequence-redundancy of the CLE domain, there is likely some specificity between pathways and/or species that are dependent on sequence. Okamoto et al. (2013) were unable to elicit a plant response in L. japonicus from exogenous application of the mature AtCLV3 peptide, but saw a reduction in nodules when LjCLE-RS2 was applied with the correct post-translational modifications. Chimeric genes that swapped the CLE domains of GmNIC1 and GmRIC1 also impacted on the suppression of nodulation compared with their respective native genes (Reid et al. 2013). In contrast, GmRIC1 overexpression in common bean and MtCLE13 overexpression in pea strongly suppressed nodulation inter-specifically (Osipova et al. 2012; Ferguson et al. 2014), indicating that these CLE peptide encoding genes can function in the AON pathways of other legume species. However, overexpression results of any kind should always be interpreted with care.
C-terminal extension domain
The C-terminal domain of the known nodulation-suppressing CLE peptides is small, at ~6–9 residues in length, and is even completely absent from some (Fig. 1; Table 1). Indeed, of the known nodulation CLE peptides, GmNIC1, PvNIC1 and MtCLE12 all lack the C-terminal extension in its entirety (Mortier et al. 2010; Reid et al. 2011a; Ferguson et al. 2014). However, the remaining nodulation CLE peptides all contain the domain, as do AtCLV3 and GmCLV3 of the CLAVATA pathway (Fiers et al. 2006; Wong et al. 2013).
The C-terminal domain is thought to act as a protective mechanism from degradative protease enzymes in the xylem, which the peptides would encounter during systemic transport (Oelkers et al. 2008; Okamoto et al. 2009; Ni et al. 2011; Reid et al. 2011a). It is characteristic of the rhizobia-dependent, systemically acting, CLE peptides, and is not present in the nitrate-induced, locally-acting GmNIC1 of soybean and its orthologue in bean, PvNIC1 (Reid et al. 2011a; Ferguson et al. 2014). This would appear to further support a role for the domain in protection during long-distance xylem transport. Moreover, overexpressing a chimeric construct that added the C-terminal domain of GmRIC1 to GmNIC1 enhanced the suppression of nodulation compared with that of the native GmNIC1 (Reid et al. 2013). In contrast, the removal of the domain from GmRIC1 did not alter its ability to suppress when overexpressed (Reid et al. 2013), but this may be due to the overexpression technique masking or over-compensating for the true function of the modified construct. MtCLE12 also lacks the C-terminal domain and is both induced by rhizobia and predicted to be transported systemically (Mortier et al. 2010), so the exact need for the domain remains puzzling.
Two conserved proline residues are present within the C-terminal extension of all seven nodulation CLE peptides that contain the domain (Fig. 1; Okamoto et al. 2009; Mortier et al. 2010; Reid et al. 2011a; Ferguson et al. 2014). Site-directed mutagenesis and overexpression of GmRIC1 modified to encode two alanine residues in place of these two proline residues did not alter the suppressive activity of the peptide, consistent with the unclear role of this domain (Reid et al. 2013).
Post-translational modifications and critical residues of the CLE domain
The mature nodulation-suppressive CLE peptide of L. japonicus, LjCLE-RS2, is 13 amino acids in length and is hydroxylated at Pro4 and Pro7, with Hyp7 further modified to contain three arabinose sugars connected via β-1-2-linkages. These modifications are predicted to be made in the extracellular fluids (Okamoto et al. 2013). This is consistent with mature AtCLV3, AtCLE2 and AtCLE9 glycopeptides, which also contain a Hyp7 having three linked L-arabinose sugars (Kondo et al. 2006; Ohyama et al. 2009; Okamoto et al. 2013; Shinohara and Matsubayashi 2013). All of the nodulation CLE peptides contain motifs associated with arabinose modifications that are present in other plant proteins/peptides (Matsubayashi 2014). The hydroxyproline O-arabinosyltransferase (HPAT) gene that controls CLE arabinosylation in Arabidopsis is called AtHPAT3 (Ogawa-Ohnishi et al. 2013). MtRDN1 and PsNOD3 are likely orthologues of AtHPAT3 and are thought to be responsible for the arabinosylation of the nodulation-suppressing CLE peptides (Ogawa-Ohnishi et al. 2013). Mutations in these genes result in a supernodulation phenotype (Jacobsen and Feenstra 1984; Postma et al. 1988; Sagan and Duc 1996; Li et al. 2009; Schnabel et al. 2011), indicating that the peptides require the arabinose sugars for their activity.
Application of synthesised arabinosylated-LjCLE-RS2 to leaves of L. japonicus plants caused a reduction in nodulation in an LjHAR1-dependent manner (Okamoto et al. 2013). However, root or shoot application of synthetic nodulation CLE peptides devoid of modifications did not affect nodulation, although altered root growth was observed (Okamoto et al. 2009; Saur et al. 2011). Moreover, application of AtCLV3 with the arabinose modifications also had no effect on nodulation (Okamoto et al. 2013). Shinohara and Matsubayashi (2013) demonstrated that the binding of the AtCLV3 CLE peptide to the AtCLV1 LRR receptor-kinase declined as the arabinose chain length decreased, whereas AtCLE9 showed no change in receptor binding efficacy to its receptor, BAM1, a CLV1/BAM-family LRR RK, in the absence of the arabinose chain (Shinohara et al. 2012). Further, tracheary element differentiation inhibitory factor (TDIF) peptides synthesised with or without hydroxyproline residues can mimic the function of the naturally occurring peptide, which contains Hyp4 and Hyp7 (Sawa et al. 2006).
In addition to post-translational modifications to critical residues, the structural configuration of the CLE peptide ligand is also likely to impact markedly on receptor interactions. Gly6 is proposed to allow for rotation, most likely because of its small size, complementing evidence for a boomerang curve in the peptide’s configuration, with both ends of the peptide bending away from the arabinosylation at Hyp7 (Okamoto et al. 2013; Shinohara and Matsubayashi 2013; Song et al. 2013). Notably, Gly6 is 100% conserved amongst the known nodulation CLE peptides (Fig. 1). Site-directed mutagenesis of Gly6 to Ala6 significantly reduced the nodule suppressive activity of GmRIC1 (Reid et al. 2013). Song et al. (2013) altered Gly6 of AtCLV3 into 18 other amino acids; no substitution was able to rescue the phenotype of Atclv3 mutant plants. Similar specificity is expected for the nodulation CLE peptides. In addition to Gly6 and Pro7, residues Arg1, Pro4, Asp8, His11 and Asn12 of GmRIC1 were required for full nodulation-suppression activity in soybean (Reid et al. 2013). Similarly, TDIF also lost activity when the CLE domain residues His1, Val3, Gly6, Asn8, Pro9 and Asn12 were changed into an alanine residue via site-directed mutagenesis (Ito et al. 2006; Sawa et al. 2006).
It has been noted that locally-acting CLE peptides, including GmNIC1 and PvNIC1 (Fig. 1), in addition to AtCLV3, GmCLV3 and LjCLV3, all contain His12 (Reid et al. 2011a; Okamoto et al. 2011; Wong et al. 2013; Ferguson et al. 2014). This may indicate a role for this residue in local, but not systemic, transport of the peptide. Constructs having swapped the CLE domain of the systemically-acting GmRIC1 and the locally-acting GmNIC1 showed an altered inhibition of nodulation when overexpressed compared with the native peptides (Reid et al. 2013). Whether residue 12 plays a specific role in the transport or recognition of the peptide is of interest to determine.
As noted above, Arg1 of the AtCLV3 CLE domain has been shown to be critical for binding and processing of the mature CLE peptide, with at least 4–5 residues upstream of Arg1 required for proper recognition of the signal (Kondo et al. 2008; Ni et al. 2011; Xu et al. 2013). It is hypothesised that a subtilisin with endoproteolytic activity cleaves the CLE peptide, with a carboxypeptidase processing the C-terminal extension where present (Ni et al. 2011; Djordjevic et al. 2011). However, to date, there is little known about the mechanisms and sites of proteolytic cleavage in the nodulation CLE peptides.
Mode of induction of the nodulation-suppressing CLE peptides
All of the known nodulation-suppressing CLE peptides are upregulated in expression by the presence of rhizobia and/or the available soil nitrogen content (Table 1). Phylogenetic analysis shows that they cluster according to their mode of induction (Fig. 2). Evidence for other environmental factors such as phosphate and soil acidity, inducing or influencing the expression of CLE peptide-encoding genes also exists.
Rhizobia-induced CLE peptides
The presence of compatible rhizobia, and possibly more specifically the rhizobia-produced Nod factor signal, elicits the expression of systemically-acting CLE peptide-encoding genes that function in AON. These CLE peptides include: LjCLE-RS1, LjCLE-RS2, MtCLE12, MtCLE13, GmRIC1, GmRIC2, PvRIC1, and PvRIC2 (Table 1; Okamoto et al. 2009, 2013; Mortier et al. 2010, 2012; Lim et al. 2011; Reid et al. 2011a, 2013; Saur et al. 2011; Hayashi et al. 2012; Ferguson et al. 2014). Overexpression of these peptides in wild-type legume plants results in a complete abolishment of nodulation, but does not alter the nodulation pattern in NARK mutants, demonstrating that they act in a NARK-dependent manner (Okamoto et al. 2009; Mortier et al. 2010; Reid et al. 2011a; Lim et al. 2011).
Laser microdissection of root sections indicate that LjCLE-RS1 and -RS2 are expressed in the stele and outside of the endodermis (cortex and epidermis) (Okamoto et al. 2009). Promoter:GUS reporter fusion studies have shown that MtCLE13 is expressed in the inner cortex during early nodulation and later in dividing cells of the cortex and pericycle. In contrast, MtCLE12 is not expressed early but instead is expressed throughout young nodules and in meristematic tissues of the elongating indeterminate nodule (Mortier et al. 2010). Finally, Lim et al. (2011) have shown that GmRIC2 is expressed in the pericycle and inner cortex during early nodule development, and later in the outer cortex of more developed nodules.
Time-course experiments have revealed different but overlapping expression patterns for these genes within a species (Table 1). Soybean GmRIC1 is induced early (within 12 h) after inoculation with infection-capable (Nod factor producing) Bradyrhizobium japonicum, whereas GmRIC2 expression is induced later (48–72 h) and remains elevated in expression for longer (Reid et al. 2011a; Hayashi et al. 2012). The rhizobia-induced peptide encoding genes of common bean, PvRIC1 and PvRIC2, exhibit a similar pattern of expression (Ferguson et al. 2014). Likewise, M. truncatula MtCLE13 is expressed earlier than MtCLE12, although both are also expressed in later stages of nodulation (Mortier et al. 2010, 2012). LjCLE-RS1 and LjCLE–RS2 are both upregulated within 3 h of inoculation (Okamoto et al. 2009). Similar to GmRIC2, MtCLE13 and LjCLE-RS1 transcript levels appear to remain elevated for longer compared with MtCLE12 and LjCLE–RS2, respectively (Okamoto et al. 2009; Mortier et al. 2010; Reid et al. 2011a).
When compared with wild-type plants, a significant increase in expression of both LjCLE-RS1 and LjCLE–RS2 was also observed in the hypernodulating mutant of L. japonicus, too much love, possibly indicating that their synthesis is directly linked to the number of nodules being formed (Magori and Kawaguchi 2010). Interestingly, the plant hormone cytokinin, which has a role in early nodule development (reviewed in Ferguson and Mathesius 2014), has also been shown to induce the expression of some nodulation-suppressing CLE peptide genes (Lim et al. 2011; Mortier et al. 2010, 2012), consistent with the idea that the initiation of the AON pathway is linked to early cell divisions. Additional studies are required to further understand the expression patterns of these nodulation-suppressing CLE peptides, both within and between species.
Nitrate-induced CLE peptides
GmNIC1 and LjCLE-RS2 are the only nodulation-suppressing CLE peptide-encoding genes that are confirmed to respond to nitrate (Table 1; Okamoto et al. 2009; Reid et al. 2011a). GmNIC1 is specifically induced by nitrate and not co-induced by the rhizobial microsymbiont, whereas LjCLE-RS2 is reported to be induced by both. PvNIC1 is also likely to be induced by nitrate as the candidate orthologue of GmNIC1 (Ferguson et al. 2014). To date, no CLE peptide-encoding gene of M. truncatula has been reported to respond to nitrate, although evidence suggests the existence of a locally acting, nitrate-responsive mechanism that acts in a MtSUNN-dependent manner to regulate nodulation (Jeudy et al. 2010). We note that AtCLE2, the Arabidopsis gene most similar to the nodulation-suppressing CLE peptides, has a role in root development and is also induced by nitrate (Scheible et al. 2004; Araya et al. 2014).
Overexpression of the locally-acting GmNIC1 in wild-type soybean reduces nodule numbers by ~50% compared with empty vector controls (Reid et al. 2011a). Although significant, this suppressive ability is far from that of GmRIC1 and GmRIC2, as discussed above. Confirmation is required to determine whether this is unique to soybean or is shared with the closely related orthologues identified in common bean (Ferguson et al. 2014).
Other inducing factors
Numerous factors can influence the extent of nodulation and it is possible that some do so by inducing, or otherwise influencing, the production, transport, perception or response to a CLE peptide(s). Recently, split-root and grafting studies using soybean grown in low pH conditions revealed a novel systemic mechanism that acts via GmNARK in the shoot to inhibit nodulation of the root (Lin et al. 2012; Ferguson et al. 2013). This suggests that soil acidity may act via a CLE peptide to suppress nodulation. Two CLE peptide-encoding genes of L. japonicus, called LjCLE19 and LjCLE20, have been shown to be upregulated in the presence of phosphate (Funayama-Noguchi et al. 2011); however, a specific role for these peptides in plant development has not been reported. It has been noted that although CLE peptides are nearly-exclusive to plants, they also exist in plant-parasitic nematodes (e.g. Bakhetia et al. 2007), which appear to use the peptides to initiate the formation of feeding structures in host roots (reviewed by Mitchum et al. 2012). Also noted is that nematodes are easily genetically transformed through simple feeding, suggesting that perhaps nematode CLE genes were plant-derived. Whether nematodes, or any other pathogen, can also induce a plant-encoded CLE peptide(s) is of great interest to determine.
Rhizobia-induced CLE peptides that do not suppress nodulation
In addition to the nodulation-suppressing CLE peptides reported above, two further CLE peptide-encoding genes have been identified that are expressed in response to rhizobia inoculation, namely LjCLE3 (Okamoto et al. 2009) and MtCLE4 (Mortier et al. 2010). Overexpression of these genes does not alter the nodulation phenotype when compared with empty vector controls. It is now of interest to determine the role of these peptides in nodulation to determine why they are responsive to rhizobia inoculation and how they function in this symbiosis.
Future perspectives
The role of CLE peptides functioning as hormone signals in plant development is only just beginning to emerge, with the activity of most remaining to be elucidated. How and where the CLE peptides are induced, whether they are transported and act locally or systemically, how they are perceived, the downstream signals they induce, and their precise role in various plant developmental pathways are all features of great interest to establish in this burgeoning research field. Indeed, establishing the function of CLE peptides acting in critical plant developmental processes will considerably help to advance the current molecular knowledgebase of a variety of plant signalling networks.
Acknowledgements
We would like to thank the Australian Research Council for provision of a Centre of Excellence grant (CEO348212) and Discovery Project grants (DP130103084 and DP130102266), as well as the Hermon Slade Foundation and the University of Queensland for strategic funds. We thank Alina Tollenaere for technical assistance in developing the phylogenetic tree in Fig. 2.
References
Araya T, Miyamoto M, Wibowo J, Suzuki A, Kojima S, Tsuchiya YN, Sawa S, Fukuda H, Von Wirén N, Takahashi H (2014) CLE-CLAVATA1 peptide-receptor signaling module regulates the expansion of plant root systems in a nitrogen-dependent manner. Proceedings of the National Academy of Sciences of the United States of America 111, 2029–2034.| CLE-CLAVATA1 peptide-receptor signaling module regulates the expansion of plant root systems in a nitrogen-dependent manner.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXitFOnt7o%3D&md5=547789e0ecf8b630638ccf11aefa6990CAS | 24449877PubMed |
Bakhetia M, Urwin PE, Atkinson HJ (2007) qPCR analysis and RNAi define pharyngeal gland cell-expressed genes of Heterodera glycines required for initial interactions with the host. Molecular Plant-Microbe Interactions 20, 306–312.
| qPCR analysis and RNAi define pharyngeal gland cell-expressed genes of Heterodera glycines required for initial interactions with the host.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXitVerurk%3D&md5=a706db2b445f71ce7fd467afa6e0ad1eCAS | 17378433PubMed |
Carroll BJ, McNeil DL, Gresshoff PM (1985a) A supernodulation and nitrate-tolerant symbiotic (nts) soybean mutant. Plant Physiology 78, 34–40.
| A supernodulation and nitrate-tolerant symbiotic (nts) soybean mutant.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaL2MXktVerur0%3D&md5=92d105d20482d7cc9dd9b9b90cdd821aCAS | 16664203PubMed |
Carroll BJ, McNeil DL, Gresshoff PM (1985b) Isolation and properties of soybean (Glycine max (L) Merr.) mutants that nodulate in the presence of high nitrate concentrations. Proceedings of the National Academy of Sciences of the United States of America 82, 4162–4166.
| Isolation and properties of soybean (Glycine max (L) Merr.) mutants that nodulate in the presence of high nitrate concentrations.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaL2MXkvVGhsLo%3D&md5=0c3811e1fbc4bc405bc980fda122a808CAS | 16593577PubMed |
Clark SE, Williams RW, Meyerowitz EM (1997) The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89, 575–585.
| The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK2sXjsVShur0%3D&md5=5071bd36bf485154fe7eb0e3214c4b76CAS | 9160749PubMed |
Cock JM, McCormick S (2001) A large family of genes that share homology with CLAVATA3. Plant Physiology 126, 939–942.
| A large family of genes that share homology with CLAVATA3.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3MXlsVartLY%3D&md5=8d7d5fabc1f77441755a3acf1f5acb73CAS | 11457943PubMed |
Delves AC, Mathews A, Day DA, Carter AS, Carroll BJ, Gresshoff PM (1986) Regulation of the soybean-Rhizobium nodule symbiosis by shoot and root factors. Plant Physiology 82, 588–590.
| Regulation of the soybean-Rhizobium nodule symbiosis by shoot and root factors.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BC3cnhslagtQ%3D%3D&md5=e378df32885efb5310b2a36f2de870cdCAS | 16665072PubMed |
Desbrosses GJ, Stougaard J (2011) Root nodulation: a paradigm for how plant-microbe symbiosis influences host developmental pathways. Cell Host & Microbe 10, 348–358.
| Root nodulation: a paradigm for how plant-microbe symbiosis influences host developmental pathways.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXhtlKitr%2FO&md5=c2f7d6232d2b103b80e1589cb23956e1CAS |
Djordjevic MA, Oakes M, Wong CE, Singh M, Bhalla P, Kusumawati L, Imin N (2011) Border sequences of Medicago truncatula CLE36 are specifically cleaved by endoproteases common to the extracellular fluids of Medicago and soybean. Journal of Experimental Botany 62, 4649–4659.
| Border sequences of Medicago truncatula CLE36 are specifically cleaved by endoproteases common to the extracellular fluids of Medicago and soybean.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXhtFKisb3O&md5=77a3eb31f781c66cee3bf8996a5a7569CAS | 21633083PubMed |
Erisman JW, Sutton MA, Galloway J, Klimont Z, Winiwarter W (2008) How a century of ammonia synthesis changed the world. Nature Geoscience 1, 636–639.
| How a century of ammonia synthesis changed the world.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXhtFOlur%2FM&md5=2a715f81a5cb1aeeadfe207c05d70b56CAS |
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791.
| Confidence limits on phylogenies: an approach using the bootstrap.Crossref | GoogleScholarGoogle Scholar |
Ferguson BJ, Mathesius U (2003) Signaling interactions during nodule development. Journal of Plant Growth Regulation 22, 47–72.
| Signaling interactions during nodule development.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3sXosF2ktbY%3D&md5=32020f2d2ee20a702dd2a5886f4b978cCAS |
Ferguson BJ, Mathesius U (2014) Phytohormone regulation of legume-rhizobia interactions. Journal of Chemical Ecology
| Phytohormone regulation of legume-rhizobia interactions.Crossref | GoogleScholarGoogle Scholar | 25052910PubMed |
Ferguson BJ, Indrasumunar A, Hayashi S, Lin M-H, Lin Y-H, Reid DE, Gresshoff PM (2010) Molecular analysis of legume nodule development and autoregulation. Journal of Integrative Plant Biology 52, 61–76.
| Molecular analysis of legume nodule development and autoregulation.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXitVOhsLo%3D&md5=c1170a0ccbd514abd34c4c6228d3618aCAS | 20074141PubMed |
Ferguson BJ, Lin MH, Gresshoff PM (2013) Regulation of legume nodulation by acidic growth conditions. Plant Signaling & Behavior 8, e23426
| Regulation of legume nodulation by acidic growth conditions.Crossref | GoogleScholarGoogle Scholar |
Ferguson BJ, Li D, Hastwell AH, Reid DE, Li Y, Jackson SA, Gresshoff PM (2014) The soybean (Glycine max) nodulation-suppressive CLE peptide, GmRIC1, functions interspecifically in common white bean (Phaseolus vulgaris), but not in a supernodulating line mutated in the receptor PvNARK. Plant Biotechnology Journal
| The soybean (Glycine max) nodulation-suppressive CLE peptide, GmRIC1, functions interspecifically in common white bean (Phaseolus vulgaris), but not in a supernodulating line mutated in the receptor PvNARK.Crossref | GoogleScholarGoogle Scholar | 25040127PubMed |
Fiers M, Golemiec E, Van Der Schors R, Van Der Geest L, Li KW, Stiekema WJ, Liu C-M (2006) The CLAVATA3/ESR motif of CLAVATA3 Is functionally independent from the nonconserved flanking sequences. Plant Physiology 141, 1284–1292.
| The CLAVATA3/ESR motif of CLAVATA3 Is functionally independent from the nonconserved flanking sequences.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XosVKitr0%3D&md5=8f432c521ca529ba428a5dc71318bc2dCAS | 16751438PubMed |
Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM (1999) Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283, 1911–1914.
| Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1MXitVantb4%3D&md5=fbe83b816fe888259f423775e6d61cfaCAS | 10082464PubMed |
Funayama-Noguchi S, Noguchi K, Yoshida C, Kawaguchi M (2011) Two CLE genes are induced by phosphate in roots of Lotus japonicus. Journal of Plant Research 124, 155–163.
| Two CLE genes are induced by phosphate in roots of Lotus japonicus.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXhsF2hu7zI&md5=3280bbc4b9f13466af0005e8a454d2a1CAS | 20428922PubMed |
Gresshoff PM, Delves AC (1986). Plant genetic approaches to symbiotic nodulation and nitrogen fixation in legumes. In ‘Plant gene research’. (Eds AD Blaustein, PJ King) pp. 159–206. (Springer-Verlag: New York)
Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology 52, 696–704.
| A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood.Crossref | GoogleScholarGoogle Scholar | 14530136PubMed |
Hayashi S, Reid DE, Lorenc MT, Stiller J, Edwards D, Gresshoff PM, Ferguson BJ (2012) Transient Nod factor-dependent gene expression in the nodulation-competent zone of soybean (Glycine max (L.) Merr.) roots. Plant Biotechnology Journal 10, 995–1010.
| Transient Nod factor-dependent gene expression in the nodulation-competent zone of soybean (Glycine max (L.) Merr.) roots.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38Xhs1art77N&md5=6ccc39fb459d6d8f5f01005e13cd9495CAS | 22863334PubMed |
Herridge D, Peoples M, Boddey R (2008) Global inputs of biological nitrogen fixation in agricultural systems. Plant and Soil 311, 1–18.
| Global inputs of biological nitrogen fixation in agricultural systems.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXhtVyju73M&md5=6a13cd768f22ac011e2df0673956f6e3CAS |
Ito Y, Nakanomyo I, Motose H, Iwamoto K, Sawa S, Dohmae N, Fukuda H (2006) Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science 313, 842–845.
| Dodeca-CLE peptides as suppressors of plant stem cell differentiation.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XnvVynt7o%3D&md5=c510662e62f55075f946ca09b29dda2cCAS | 16902140PubMed |
Jacobsen E, Feenstra WJ (1984) A new pea mutant with efficient nodulation in the presence of nitrate. Plant Science Letters 33, 337–344.
| A new pea mutant with efficient nodulation in the presence of nitrate.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaL2cXhvVyqtrw%3D&md5=4bc44d74507877f31219a1e9f46c0bebCAS |
Jensen E, Peoples M, Boddey R, Gresshoff P, Hauggaard-Nielsen H, Alves B, Morrison M (2012) Legumes for mitigation of climate change and feedstock in a bio-based economy – a review. Agronomy for Sustainable Development 32, 329–364.
| Legumes for mitigation of climate change and feedstock in a bio-based economy – a review.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XksVOgt78%3D&md5=9a5f573d6c29cf5999ba2b8b3cd86f70CAS |
Jeudy C, Ruffel S, Freixes S, Tillard P, Santoni AL, Morel S, Journet E-P, Duc G, Gojon A, Lepetit M, Salon C (2010) Adaptation of Medicago truncatula to nitrogen limitation is modulated via local and systemic nodule developmental responses. New Phytologist 185, 817–828.
| Adaptation of Medicago truncatula to nitrogen limitation is modulated via local and systemic nodule developmental responses.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXitFKns7k%3D&md5=420d5f942614eac9f1981fb403e1114bCAS | 20015066PubMed |
Kondo T, Sawa S, Kinoshita A, Mizuno S, Kakimoto T, Fukuda H, Sakagami Y (2006) A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science 313, 845–848.
| A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XnvVynt7s%3D&md5=0a3fa4726970055b586a4ec595538ce8CAS | 16902141PubMed |
Kondo T, Nakamura T, Yokomine K, Sakagami Y (2008) Dual assay for MCLV3 activity reveals structure-activity relationship of CLE peptides. Biochemical and Biophysical Research Communications 377, 312–316.
| Dual assay for MCLV3 activity reveals structure-activity relationship of CLE peptides.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXhtlaqsb3L&md5=4580637ce158e4e73951136d0c7c7619CAS | 18848920PubMed |
Kosslak RM, Bohlool BB (1984) Suppression of nodule development of one side of a split-root system of soybeans caused by prior inoculation of the other side. Plant Physiology 75, 125–130.
| Suppression of nodule development of one side of a split-root system of soybeans caused by prior inoculation of the other side.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BC3cnhsFOhtA%3D%3D&md5=dc4a52cba455da6bdab7f9538ea48cadCAS | 16663555PubMed |
Krusell L, Madsen LH, Sato S, Aubert G, Genua A, Szczyglowski K, Duc G, Kaneko T, Tabata S, De Bruijn F, Pajuelo E, Sandal N, Stougaard J (2002) Shoot control of root development and nodulation is mediated by a receptor-like kinase. Nature 420, 422–426.
| Shoot control of root development and nodulation is mediated by a receptor-like kinase.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38XptFCis7w%3D&md5=6c61bafbfcba361b0cdf1919632019f1CAS | 12442170PubMed |
Krusell L, Sato N, Fukuhara I, Koch BEV, Grossmann C, Okamoto S, Oka-Kira E, Otsubo Y, Aubert G, Nakagawa T, Sato S, Tabata S, Duc G, Parniske M, Wang TL, Kawaguchi M, Stougaard J (2011) The Clavata2 genes of pea and Lotus japonicus affect autoregulation of nodulation. The Plant Journal 65, 861–871.
| The Clavata2 genes of pea and Lotus japonicus affect autoregulation of nodulation.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXkvVais7c%3D&md5=683c6555ea0afe96eb6a940b5ea84ca2CAS | 21276104PubMed |
la Cour T, Kiemer L, Mølgaard A, Gupta R, Skriver K, Brunak S (2004) Analysis and prediction of leucine-rich nuclear export signals. Protein Engineering, Design & Selection 17, 527–536.
| Analysis and prediction of leucine-rich nuclear export signals.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXos1GisLc%3D&md5=c6523ad8d9eed346057e1b3851931465CAS |
Larkin MA, Blackshields G, Brown NP, Chenna R, Mcgettigan PA, Mcwilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948.
| Clustal W and Clustal X version 2.0.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXhtlaqsL%2FM&md5=a7e5427f5948a5538b27ba99db4e46c4CAS | 17846036PubMed |
Li D, Kinkema M, Gresshoff PM (2009) Autoregulation of nodulation (AON) in Pisum sativum (pea) involves signalling events associated with both nodule primordia development and nitrogen fixation. Journal of Plant Physiology 166, 955–967.
| Autoregulation of nodulation (AON) in Pisum sativum (pea) involves signalling events associated with both nodule primordia development and nitrogen fixation.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXmslarur8%3D&md5=5602d512011b6d459a8b867b9904edd5CAS | 19403196PubMed |
Lim CW, Lee YW, Hwang CH (2011) Soybean nodule-enhanced CLE peptides in roots act as signals in GmNARK-mediated nodulation suppression. Plant & Cell Physiology 52, 1613–1627.
| Soybean nodule-enhanced CLE peptides in roots act as signals in GmNARK-mediated nodulation suppression.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXht1SiurrJ&md5=377c570f003f5cc6b3c3e2fa6d648d15CAS |
Lin Y-H, Ferguson BJ, Kereszt A, Gresshoff PM (2010) Suppression of hypernodulation in soybean by a leaf-extracted, NARK- and Nod factor-dependent, low molecular mass fraction. New Phytologist 185, 1074–1086.
| Suppression of hypernodulation in soybean by a leaf-extracted, NARK- and Nod factor-dependent, low molecular mass fraction.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXksFCrt7Y%3D&md5=697580f860a786edd0dc9e3a563365adCAS | 20100211PubMed |
Lin Y-H, Lin M-H, Gresshoff PM, Ferguson BJ (2011) An efficient petiole-feeding bioassay for introducing aqueous solutions into dicotyledonous plants. Nature Protocols 6, 36–45.
| An efficient petiole-feeding bioassay for introducing aqueous solutions into dicotyledonous plants.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXisVShsQ%3D%3D&md5=1dec4c966e6a9a4feb1756ee1e26f25cCAS | 21212781PubMed |
Lin M-H, Gresshoff PM, Ferguson BJ (2012) Systemic regulation of soybean nodulation by acidic growth conditions. Plant Physiology 160, 2028–2039.
| Systemic regulation of soybean nodulation by acidic growth conditions.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XhvVKmt7fM&md5=0f9a68c07585bcb3eccd60edd2455aeaCAS | 23054568PubMed |
Magori S, Kawaguchi M (2010) Analysis of two potential long-distance signaling molecules, LjCLE-RS1/2and jasmonic acid, in a hypernodulating mutant too much love. Plant Signaling & Behavior 5, 403–405.
| Analysis of two potential long-distance signaling molecules, LjCLE-RS1/2and jasmonic acid, in a hypernodulating mutant too much love.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXhsFemu73K&md5=7432168155ca38fec35e9b55f6336319CAS |
Magori S, Oka-Kira E, Shibata S, Umehara Y, Kouchi H, Hase Y, Tanaka A, Sato S, Tabata S, Kawaguchi M (2009) TOO MUCH LOVE, a root regulator associated with the long-distance control of nodulation in Lotus japonicus. Molecular Plant-Microbe Interactions 22, 259–268.
| TOO MUCH LOVE, a root regulator associated with the long-distance control of nodulation in Lotus japonicus.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXisVWmsrs%3D&md5=dc2d472db13d6aa3d22308edfdde73b8CAS | 19245320PubMed |
Matsubayashi Y (2014) Posttranslationally modified small-peptide signals in plants. Annual Review of Plant Biology 65, 385–413.
| Posttranslationally modified small-peptide signals in plants.Crossref | GoogleScholarGoogle Scholar | 24779997PubMed |
Meng L, Ruth KC, Fletcher JC, Feldman L (2010) The roles of different CLE domains in Arabidopsis CLE polypeptide activity and functional specificity. Molecular Plant 3, 760–772.
| The roles of different CLE domains in Arabidopsis CLE polypeptide activity and functional specificity.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXpsFKktrY%3D&md5=4f87ced4a1173961fe6122ca47c1509dCAS | 20494950PubMed |
Mitchum MG, Wang X, Wang J, Davis EL (2012) Role of nematode peptides and other small molecules in plant parasitism. Annual Review of Phytopathology 50, 175–195.
| Role of nematode peptides and other small molecules in plant parasitism.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XhsVWht7zE&md5=f3f224515cc3f580ad672d8186a81afdCAS | 22578179PubMed |
Miyahara A, Hirani TA, Oakes M, Kereszt A, Kobe B, Djordjevic MA, Gresshoff PM (2008) Soybean nodule autoregulation receptor kinase phosphorylates two kinase-associated protein phosphatases in vitro. The Journal of Biological Chemistry 283, 25381–25391.
| Soybean nodule autoregulation receptor kinase phosphorylates two kinase-associated protein phosphatases in vitro.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXhtVyisLvK&md5=b985927ceffd766be6dcb8dbb709d9d4CAS | 18606823PubMed |
Miyazawa H, Oka-Kira E, Sato N, Takahashi H, Wu G-J, Sato S, Hayashi M, Betsuyaku S, Nakazono M, Tabata S, Harada K, Sawa S, Fukuda H, Kawaguchi M (2010) The receptor-like kinase KLAVIER mediates systemic regulation of nodulation and non-symbiotic shoot development in Lotus japonicus. Development 137, 4317–4325.
| The receptor-like kinase KLAVIER mediates systemic regulation of nodulation and non-symbiotic shoot development in Lotus japonicus.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXhtFGmurk%3D&md5=65ebf69aea34e0340cb41677663276e9CAS | 21098572PubMed |
Mortier V, Den Herder G, Whitford R, Van De Velde W, Rombauts S, D’haeseleer K, Holsters M, Goormachtig S (2010) CLE peptides control Medicago truncatula nodulation locally and systemically. Plant Physiology 153, 222–237.
| CLE peptides control Medicago truncatula nodulation locally and systemically.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXnt1Ggs74%3D&md5=514c973fce7405edd87e8c4421490bd2CAS | 20348212PubMed |
Mortier V, De Wever E, Vuylsteke M, Holsters M, Goormachtig S (2012) Nodule numbers are governed by interaction between CLE peptides and cytokinin signaling. The Plant Journal 70, 367–376.
| Nodule numbers are governed by interaction between CLE peptides and cytokinin signaling.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XmslOgtbc%3D&md5=33e60135fc842f3acbe46814fdf17c12CAS | 22168914PubMed |
Ni J, Clark SE (2006) Evidence for functional conservation, sufficiency, and proteolytic processing of the CLAVATA3 CLE domain. Plant Physiology 140, 726–733.
| Evidence for functional conservation, sufficiency, and proteolytic processing of the CLAVATA3 CLE domain.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XjsV2iu7w%3D&md5=23d058ce2b5a3d85f9bba7a3e2ca4710CAS | 16407446PubMed |
Ni J, Guo Y, Jin H, Hartsell J, Clark S (2011) Characterization of a CLE processing activity. Plant Molecular Biology 75, 67–75.
| Characterization of a CLE processing activity.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXhsF2gsbnF&md5=4b4d1bd9e3e1571c014a47e217a858e5CAS | 21052783PubMed |
Nishimura R, Hayashi M, Wu G-J, Kouchi H, Imaizumi-Anraku H, Murakami Y, Kawasaki S, Akao S, Ohmori M, Nagasawa M, Harada K, Kawaguchi M (2002) HAR1 mediates systemic regulation of symbiotic organ development. Nature 420, 426–429.
| HAR1 mediates systemic regulation of symbiotic organ development.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38XptFCis7k%3D&md5=07ae29a28244fd52d5f1ca34101c4068CAS | 12442172PubMed |
Nontachaiyapoom S, Scott PT, Men AE, Kinkema M, Schenk PM, Gresshoff PM (2007) Promoters of orthologous Glycine max and Lotus japonicus nodulation autoregulation genes interchangeably drive phloem-specific expression in transgenic plants. Molecular Plant-Microbe Interactions 20, 769–780.
| Promoters of orthologous Glycine max and Lotus japonicus nodulation autoregulation genes interchangeably drive phloem-specific expression in transgenic plants.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXmvFanurY%3D&md5=77ed933de9383d50cba910c4c095379fCAS | 17601165PubMed |
Oelkers K, Goffard N, Weiller G, Gresshoff PM, Mathesius U, Frickey T (2008) Bioinformatic analysis of the CLE signaling peptide family. BMC Plant Biology 8, 1
| Bioinformatic analysis of the CLE signaling peptide family.Crossref | GoogleScholarGoogle Scholar | 18171480PubMed |
Ogawa M, Shinohara H, Sakagami Y, Matsubayashi Y (2008) Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science 319, 294
| Arabidopsis CLV3 peptide directly binds CLV1 ectodomain.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXmt1Oqtw%3D%3D&md5=127310ceaa9216700317576513c96946CAS | 18202283PubMed |
Ogawa-Ohnishi M, Matsushita W, Matsubayashi Y (2013) Identification of three hydroxyproline O-arabinosyltransferases in Arabidopsis thaliana. Nature Chemical Biology 9, 726–730.
| Identification of three hydroxyproline O-arabinosyltransferases in Arabidopsis thaliana.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXhsVeru7fP&md5=52ae06719c4c648dd01f85d48064f7fdCAS | 24036508PubMed |
Ohyama K, Shinohara H, Ogawa-Ohnishi M, Matsubayashi Y (2009) A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nature Chemical Biology 5, 578–580.
| A glycopeptide regulating stem cell fate in Arabidopsis thaliana.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXnt1Sht70%3D&md5=fa47971b8db75f9700a20469068b1c9bCAS | 19525968PubMed |
Okamoto S, Ohnishi E, Sato S, Takahashi H, Nakazono M, Tabata S, Kawaguchi M (2009) Nod Factor/nitrate-induced CLE genes that drive HAR1-mediated systemic regulation of nodulation. Plant & Cell Physiology 50, 67–77.
| Nod Factor/nitrate-induced CLE genes that drive HAR1-mediated systemic regulation of nodulation.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXhtVegsrw%3D&md5=129484f4016e20269424aa4e497341abCAS |
Okamoto S, Nakagawa T, Kawaguchi M (2011) Expression and functional analysis of a CLV3-like gene in the model legume Lotus japonicus. Plant & Cell Physiology 52, 1211–1221.
| Expression and functional analysis of a CLV3-like gene in the model legume Lotus japonicus.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXoslOisrY%3D&md5=6c4808f773633a90c73ba966a2049491CAS |
Okamoto S, Shinohara H, Mori T, Matsubayashi Y, Kawaguchi M (2013) Root-derived CLE glycopeptides control nodulation by direct binding to HAR1 receptor kinase. Nature Communications 4, 2191
| Root-derived CLE glycopeptides control nodulation by direct binding to HAR1 receptor kinase.Crossref | GoogleScholarGoogle Scholar | 23934307PubMed |
Oldroyd GED (2013) Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nature Reviews. Microbiology 11, 252–263.
| Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXktVKjtLc%3D&md5=8f6924e546ce67182911fa224e351fe3CAS |
Osipova MA, Mortier V, Demchenko KN, Tsyganov VE, Tikhonovich IA, Lutova LA, Dolgikh EA, Goormachtig S (2012) WUSCHEL-RELATED HOMEOBOX5 gene expression and interaction of CLE peptides with components of the systemic control add two pieces to the puzzle of autoregulation of nodulation. Plant Physiology 158, 1329–1341.
| WUSCHEL-RELATED HOMEOBOX5 gene expression and interaction of CLE peptides with components of the systemic control add two pieces to the puzzle of autoregulation of nodulation.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XltVOksbg%3D&md5=8a9687ddced2eed4c196d19ea47cb53aCAS | 22232385PubMed |
Postma JG, Jacobsen E, Feenstra WJ (1988) Three pea mutants with an altered nodulation studied by genetic analysis and grafting. Journal of Plant Physiology 132, 424–430.
| Three pea mutants with an altered nodulation studied by genetic analysis and grafting.Crossref | GoogleScholarGoogle Scholar |
Reid DE, Ferguson BJ, Gresshoff PM (2011a) Inoculation- and nitrate-induced CLE peptides of soybean control NARK-dependent nodule formation. Molecular Plant-Microbe Interactions 24, 606–618.
| Inoculation- and nitrate-induced CLE peptides of soybean control NARK-dependent nodule formation.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXltF2ltrY%3D&md5=796db89694ae285141ac0ca2fbdfd87dCAS | 21198362PubMed |
Reid DE, Ferguson BJ, Hayashi S, Lin Y-H, Gresshoff PM (2011b) Molecular mechanisms controlling legume autoregulation of nodulation. Annals of Botany 108, 789–795.
| Molecular mechanisms controlling legume autoregulation of nodulation.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXht1eisbrE&md5=f360a37ca6bbfde308aa7a4680cb30deCAS | 21856632PubMed |
Reid DE, Hayashi S, Lorenc M, Stiller J, Edwards D, Gresshoff PM, Ferguson BJ (2012) Identification of systemic responses in soybean nodulation by xylem sap feeding and complete transcriptome sequencing reveal a novel component of the autoregulation pathway. Plant Biotechnology Journal 10, 680–689.
| Identification of systemic responses in soybean nodulation by xylem sap feeding and complete transcriptome sequencing reveal a novel component of the autoregulation pathway.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XhsVWgurfK&md5=d9722f2710bbb1e31558a3cf6f53bc93CAS | 22624681PubMed |
Reid DE, Li D, Ferguson BJ, Gresshoff PM (2013) Structure–function analysis of the GmRIC1 signal peptide and CLE domain required for nodulation control in soybean. Journal of Experimental Botany 64, 1575–1585.
| Structure–function analysis of the GmRIC1 signal peptide and CLE domain required for nodulation control in soybean.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXlsV2ktbY%3D&md5=9a4a13f836d326788599a7c243e3714eCAS | 23386683PubMed |
Rojo E, Sharma V, Kovaleva V, Raikhel N, Fletcher J (2002) CLV3 is localized to the extracellular space, where it activates the Arabidopsis CLAVATA stem cell signaling pathway. The Plant Cell 14, 969–977.
| CLV3 is localized to the extracellular space, where it activates the Arabidopsis CLAVATA stem cell signaling pathway.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38XksVGqsbs%3D&md5=bb4c65c508218fbd6aebf9a12602eb39CAS | 12034890PubMed |
Sagan M, Duc G (1996) Sym28 and Sym29, two new genes involved in regulation of nodulation in pea (Pisum sativum L.). Symbiosis 20, 229–245.
Sasaki T, Suzaki T, Soyano T, Kojima M, Sakakibara H, Kawaguchi M (2014) Shoot-derived cytokinins systemically regulate root nodulation. Nature Communications 5, 4983
| Shoot-derived cytokinins systemically regulate root nodulation.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXitVWgsLnM&md5=5792682d9bdb8d7e1caae93095970362CAS | 25236855PubMed |
Saur IML, Oakes M, Djordjevic MA, Imin N (2011) Crosstalk between the nodulation signaling pathway and the autoregulation of nodulation in Medicago truncatula. New Phytologist 190, 865–874.
| Crosstalk between the nodulation signaling pathway and the autoregulation of nodulation in Medicago truncatula.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXot1Crur4%3D&md5=b94ea9cdc1037da750cf875fdbb29336CAS |
Sawa S, Kinoshita A, Nakanomyo I, Fukuda H (2006) CLV3/ESR-related (CLE) peptides as intercellular signaling molecules in plants. Chemical Record 6, 303–310.
| CLV3/ESR-related (CLE) peptides as intercellular signaling molecules in plants.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXksVWmtLk%3D&md5=bb339d0d4003ec787a8baba735fbe019CAS | 17304552PubMed |
Scheible W-R, Morcuende R, Czechowski T, Fritz C, Osuna D, Palacios-Rojas N, Schindelasch D, Thimm O, Udvardi MK, Stitt M (2004) Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiology 136, 2483–2499.
| Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXnvFOrtbc%3D&md5=bfd116c6503b4f1dc68a09764f2ce255CAS | 15375205PubMed |
Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, Xu D, Hellsten U, May GD, Yu Y, Sakurai T, Umezawa T, Bhattacharyya MK, Sandhu D, Valliyodan B, Lindquist E, Peto M, Grant D, Shu S, Goodstein D, Barry K, Futrell-Griggs M, Abernathy B, Du J, Tian Z, Zhu L, Gill N, Joshi T, Libault M, Sethuraman A, Zhang X-C, Shinozaki K, Nguyen HT, Wing RA, Cregan P, Specht J, Grimwood J, Rokhsar D, Stacey G, Shoemaker RC, Jackson SA (2010) Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183.
| Genome sequence of the palaeopolyploid soybean.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXntVClsQ%3D%3D&md5=26223eabf9b254cb7bb9c6268bb834d2CAS | 20075913PubMed |
Schnabel E, Journet E-P, Carvalho-Niebel DF, Duc G, Frugoli J (2005) The Medicago truncatula SUNN gene encodes a CLV1-like Leucine-rich repeat receptor kinase that regulates nodule number and root length. Plant Molecular Biology 58, 809–822.
| The Medicago truncatula SUNN gene encodes a CLV1-like Leucine-rich repeat receptor kinase that regulates nodule number and root length.Crossref | GoogleScholarGoogle Scholar | 16240175PubMed |
Schnabel E, Kassaw T, Smith L, Marsh J, Oldroyd GE, Long SR, Frugoli J (2011) ROOT DETERMINED NODULATION 1 regulates nodule number in M. truncatula and defines a highly conserved, uncharacterized plant gene family. Plant Physiology 157, 328–340.
| ROOT DETERMINED NODULATION 1 regulates nodule number in M. truncatula and defines a highly conserved, uncharacterized plant gene family.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXht1Sit7zK&md5=ebc25bd3485d08abf3fd1642f0c207d0CAS | 21742814PubMed |
Searle IR, Men AE, Laniya TS, Buzas DM, Iturbe-Ormaetxe I, Carroll BJ, Gresshoff PM (2003) Long-distance signaling in nodulation directed by a CLAVATA1-like receptor kinase. Science 299, 109–112.
| Long-distance signaling in nodulation directed by a CLAVATA1-like receptor kinase.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38XpvVensLo%3D&md5=e01908423ff2786a8e2ea992db1003d7CAS | 12411574PubMed |
Shinohara H, Matsubayashi Y (2013) Chemical synthesis of Arabidopsis CLV3 glycopeptide reveals the impact of hydroxyproline arabinosylation on peptide conformation and activity. Plant & Cell Physiology 54, 369–374.
| Chemical synthesis of Arabidopsis CLV3 glycopeptide reveals the impact of hydroxyproline arabinosylation on peptide conformation and activity.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXjslGmtrc%3D&md5=721ae47a09fd6bc992aefd84bdbe417cCAS |
Shinohara H, Moriyama Y, Ohyama K, Matsubayashi Y (2012) Biochemical mapping of a ligand-binding domain within Arabidopsis BAM1 reveals diversified ligand recognition mechanisms of plant LRR-RKs. The Plant Journal 70, 845–854.
| Biochemical mapping of a ligand-binding domain within Arabidopsis BAM1 reveals diversified ligand recognition mechanisms of plant LRR-RKs.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38Xps12qsb8%3D&md5=5b007856dc0097381e5c7dc2ab8be465CAS | 22321211PubMed |
Song X-F, Guo P, Ren S-C, Xu T-T, Liu C-M (2013) Antagonistic peptide technology for functional dissection of CLV3/ESR genes in Arabidopsis. Plant Physiology 161, 1076–1085.
| Antagonistic peptide technology for functional dissection of CLV3/ESR genes in Arabidopsis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXmvFKrtLw%3D&md5=719d21416e409855d90ca9861ea65be9CAS | 23321419PubMed |
Soyano T, Hirakawa H, Sato S, Hayashi M, Kawaguchi M (2014) NODULE INCEPTION creates a long-distance negative feedback loop involved in homeostatic regulation of nodule organ production. Proceedings of the National Academy of Sciences of the United States of America
| NODULE INCEPTION creates a long-distance negative feedback loop involved in homeostatic regulation of nodule organ production.Crossref | GoogleScholarGoogle Scholar | 25246578PubMed |
Stefanović S, Pfeil BE, Palmer JD, Doyle JJ (2009) Relationships among phaseoloid legumes based on sequences from eight chloroplast regions. Systematic Botany 34, 115–128.
| Relationships among phaseoloid legumes based on sequences from eight chloroplast regions.Crossref | GoogleScholarGoogle Scholar |
Strabala TJ, O’donnell PJ, Smit A-M, Ampomah-Dwamena C, Martin EJ, Netzler N, Nieuwenhuizen NJ, Quinn BD, Foote HCC, Hudson KR (2006) Gain-of-function phenotypes of many CLAVATA3/ESR genes, including four new family members, correlate with tandem variations in the conserved CLAVATA3/ESR domain. Plant Physiology 140, 1331–1344.
| Gain-of-function phenotypes of many CLAVATA3/ESR genes, including four new family members, correlate with tandem variations in the conserved CLAVATA3/ESR domain.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28Xjslyqu7k%3D&md5=b5145f611c9bb80190aab6073a0cc030CAS | 16489133PubMed |
Sutton MA, Oenema O, Erisman JW, Leip A, Van Grinsven H, Winiwarter W (2011) Too much of a good thing. Nature 472, 159–161.
| Too much of a good thing.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXksFWgt74%3D&md5=c81b5195ad410c8f2fa1cf777e374997CAS | 21478874PubMed |
Takahara M, Magori S, Soyano T, Okamoto S, Yoshida C, Yano K, Sato S, Tabata S, Yamaguchi K, Shigenobu S, Takeda N, Suzaki T, Kawaguchi M (2013) TOO MUCH LOVE, a novel kelch repeat-containing F-box protein, functions in the long-distance regulation of the legume–Rhizobium symbiosis. Plant & Cell Physiology 54, 433–447.
| TOO MUCH LOVE, a novel kelch repeat-containing F-box protein, functions in the long-distance regulation of the legume–Rhizobium symbiosis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXlsV2mtr0%3D&md5=2212f376cfdfb8da29547df2b0a357abCAS |
Udvardi MK, Price GD, Gresshoff PM, Day DA (1988) A dicarboxylate transporter on the peribacteroid membrane of soybean nodules. FEBS Letters 231, 36–40.
| A dicarboxylate transporter on the peribacteroid membrane of soybean nodules.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaL1cXktlGnsbs%3D&md5=a77244f56509314506220bd335eca254CAS |
Wong CE, Singh MB, Bhalla PL (2013) Spatial expression of CLAVATA3 in the shoot apical meristem suggests it is not a stem cell marker in soybean. Journal of Experimental Botany 64, 5641–5649.
| Spatial expression of CLAVATA3 in the shoot apical meristem suggests it is not a stem cell marker in soybean.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXotlOn&md5=9d0d2ecbf52ea61d6f1c37b948fff4f3CAS | 24179098PubMed |
Xu T-T, Song X-F, Ren S-C, Liu C-M (2013) The sequence flanking the N-terminus of the CLV3 peptide is critical for its cleavage and activity in stem cell regulation in Arabidopsis. BMC Plant Biology 13, 225
| The sequence flanking the N-terminus of the CLV3 peptide is critical for its cleavage and activity in stem cell regulation in Arabidopsis.Crossref | GoogleScholarGoogle Scholar | 24369789PubMed |


