Evolutionary advantages of secreted peptide signalling molecules in plants
Janet I. Wheeler A and Helen R. Irving A BA Monash Institute of Pharmaceutical Sciences, Monash University, 381 Royal Parade, Parkville, Vic. 3052, Australia.
B Corresponding author. Email: helen.irving@pharm.monash.edu.au
This paper is part of an ongoing series: ‘The Evolution of Plant Functions’.
Functional Plant Biology 37(5) 382-394 https://doi.org/10.1071/FP09242
Submitted: 2 October 2009 Accepted: 2 February 2010 Published: 30 April 2010
Abstract
Peptide signalling molecules create diverse modular signals in animal systems, but it is only relatively recently that an expanding array of peptide signalling groups has been identified in plants. Representatives occur in moss although most are in angiosperms (both monocot and dicot) including many agronomically important crops. Some groups show high diversity within a species, whereas other peptide signalling groups are small or represented by a single peptide or only found in a single family of plants. Plant peptide signals regulate meristem organogenesis and growth, modulate plant homeostasis and growth, and recognise damage or imminent danger from pathogen attack. The peptide signalling molecules are secreted into the apoplast where they are often further proteolytically processed before acting on receptors in nearby or adjacent cells with all the hallmarks of paracrine molecules. Where the receptors have been identified, they are receptor-like kinases that form oligomers upon peptide binding and relay messages via phosphorylation cascades. The use of nitrogen rich amino acids in the signalling peptides was analysed and nitrogen scores were obtained that are higher than the mean nitrogen score for the overall average of the Arabidopsis proteome. These findings are discussed in terms of nutritional availability and energy use.
Additional keywords: Clavata3 (CLV3), CLE peptides, C-terminally encoded peptide 1 (CEP1), Embryo Surrounding Region (ESR), Epidermal Patterning Factor (EPF), Inflorescence Deficient in Abscission (IDA), leucine-rich repeat receptor-like kinases (LRR-RLKs), PEP1 peptide, phytosulfokine (PSK), plant natriuretic peptide (PNP), Rapid Alkalisation Factor (RALF), S-locus cysteine rich (SCR) proteins, Tapetum Determinant1 (TPD1).
Introduction
Plants are highly complex sessile living organisms that have evolved many methods to respond rapidly to environmental changes to continue normal growth and development. This is, in part, achieved by complex signalling processes mediated through networks of regulatory proteins and hormones. Peptide signalling molecules create diverse modular signals in animal systems, but it is only relatively recently that this class of molecules has been recognised in plants. In the last 20 years, peptide signalling molecules have been shown to contribute to a wide variety of plant functions ranging from plant cell differentiation to host defence responses (some recent reviews include Farrokhi et al. 2008; Jun et al. 2008; Boller and Felix 2009; Butenko et al. 2009). This is in addition to the arsenal of plant defence peptides and proteins with anti-microbial activity or the many protease inhibitors with Arabidopsis thaliana (L.) alone having 41 proteinase inhibitors (for a review, see Farrokhi et al. 2008).
In this review, we examine peptide molecules that are secreted and act in the extracellular apoplastic space to regulate plant growth, development, defence and other stress responses. We focus on secreted peptide signalling molecules found across a range of species including A. thaliana. Many of these molecules are listed in Table 1 and a brief review on each peptide family is available in the Accessory Publication to this paper. In this report, we briefly review the role of the various peptide signalling molecule classes in development and stress responses, and discuss some generalised themes that have become evident from combinations of biochemical, genetic and molecular biology studies over recent years. Finally, we use peptide nitrogen and sulfur content analysis as a way to assess the importance of these peptides to the plant proteome where we argue that the peptide molecules form an energy efficient means to allow gradients of signalling molecules to occur in niche areas within the plant.

|
Systemin: the discovery of peptide signalling systems
The first peptide signalling molecule identified was systemin, which was isolated from wounded tomato leaves (Solanum lycopersicum L.), where it induces synthesis of proteinase inhibitors (Pearce et al. 1991). Systemin is an 18 amino acid peptide product processed from the C-terminal of prosystemin, a 200 amino acid precursor protein (Dombrowski et al. 1999). Along with systemin, jasmonic acid (JA) has also been associated with early wound response (Farmer et al. 1992). In their review, Schilmiller and Howe (2005) describe grafting experiments in tomato, which show that both systemin and JA synthesis are required in the wounded tissue for a systemic response (high levels of proteinase inhibitors in upper leaves) and that this wound response required leaves to perceive but not synthesise jasmonate. The overexpression of prosystemin leads to expression of a systemic signal in tomato that induces the systemic response (McGurl et al. 1994). Although homologues of systemin have been found in other species such as potato (Solanum tuberosum L.), bell pepper (Capsicum annuum L.) and black nightshade (Solanum nigrum L.), it is restricted to species within the Solaneae subtribe (Ryan and Pearce 1998), which is suggestive of systemin developing after the divergence of Nicotiana and Solanum.
Surprisingly, systemin was shown to bind to SR160, which is the tomato homologue of AtBRI1, the brassinosteroid receptor (Scheer and Ryan 2002). AtBRI1 is a leucine-rich repeat receptor-like kinase (LRR-RLK) that activates a well characterised phosphorylation cascade beginning with receptor autophosphorylation in response to brassinosteroids (Wang et al. 2005, 2008). Subsequently, the brassinosteroid mutant cu3 was found to be as sensitive to systemin as wild-type plants (Holton et al. 2007; Lanfermeijer et al. 2008). These conflicting results were recently clarified by Malinowski et al. (2009), who showed that while systemin does bind specifically to SR160, systemin does not activate the BRI1 receptor autophosphorylation cascade. However, the characteristic signalling responses of the systemin pathway are induced, indicating that the true systemin receptor is still to be identified. In addition, there was also no difference in the level of proteinase inhibitors between tomato plants that overexpressed prosystemin and those that had the SR160 silenced (Malinowski et al. 2009). These findings highlight the importance of using binding and phosphorylation assays in addition to combinations of knock-down interactions to determine the veracity of the receptor–ligand complex.
Phylogenic relationships between classes
Most of the peptide groups examined have representatives in agronomically important monocot and dicot lineages such as rice (Oryza sativa L.), maize (Zea mays L.), sorghum (Sorghum bicolour (L.) Moench), soybean (Glycine max (L.) Merr.), castor oil bean (Ricinus communic L.), wine grape (Vitis Vinifera L.) and the black cottonwood tree (Populus balsamifera L. ssp. trichocarpa (Torr. & A. Gray ex hook.) Brayshaw). Some peptide groups such as plant peptide containing sulfated tyrosine (PSY) and S-locus cysteine-rich-like (SCRL) are similar to systemin in that representatives have been found in fewer species. A smaller subset of peptide groups are also represented in the conifers namely: Rapid Alkalinisation Factor (RALF), Phytosulfokine (PSK) and Epidermal Patterning Factor (EPF). Plant Natriuretic Peptide (PNP; sometimes annotated as expansin-like), Tapetum Determinant (TPD) and EPF are found in moss, and the conserved six cysteine residues within the carboxy terminal of EPF peptides are also found in sea anemone sequences.
Closer examination of the 126 A. thaliana peptide sequences available shows that peptides of the same group are more similar to each other than any peptide from another group (see Fig. S1 available as an Accessory Publication to this paper). A single protein sequence was selected to represent each peptide group and a radial cobalt tree shows the peptides in relation to one another (Fig. 1) where, for instance, Clavata3 (CLV3) was chosen to represent the CLE family. CLV3 is the prototype member of the CLE family named after Clv3 from A. thaliana (Fletcher et al. 1999) and Embryo Surrounding Region (ESR) from maize (Opsahl-Ferstad et al. 1997) and forms one of the largest families of plant peptide signalling molecules present throughout the plant kingdom (Cock and McCormack 2001; Oelkers et al. 2008), with over 30 annotated genes in A. thaliana.
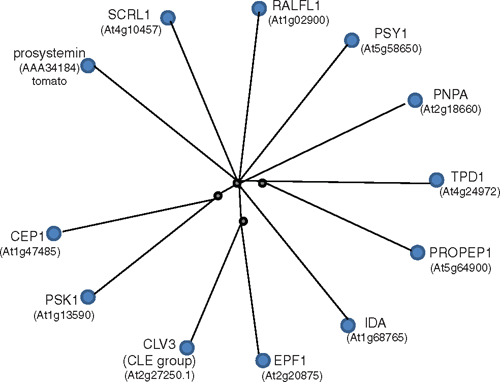
|
The number of members of the peptide groups we examined varied from 1 (TPD1) to 34 (RALF and RAFL-like (RALFL)), and groups with many members did not necessarily correlate with ancient lineages. A TPD1 sequence homologue has been found in moss but the prolific SCRL family has 26 members in A. thaliana but is limited to the Brassicaceae (Schopfer et al. 1999). Within each peptide group, the members were often spread across the chromosomes but in some cases, the peptide-encoding genes were clustered (see Fig. S1). This is seen with PROPEP (the full length forms of the peptide signalling family involved in innate defence responses) where six genes are clustered in two groups on chromosome 5, and another gene (PROPEP6) occurs on chromosome 2, which is more similar to members of one of the clusters than the other. Similarly, the more expanded signalling peptide groups such as RALFL, SCRL and CLE also have clustered encoding genes. In the case of RALFL, several encoding gene groups are clustered and are very similar in sequence, suggesting recent duplication such as RALFL2 with RALFL3 and RALFL8 with RALFL9. However, close proximity of encoding genes does not necessarily mean there is a high level of sequence similarity (see Fig. S1).
Processing of peptide signalling molecules
In general, peptide signalling molecules are relatively small proteins (ranging in size from ~60 to 180 amino acids) containing a N-terminal secretory signal sequence (pre-propeptide) that is cleaved off by endoplasmic reticulum proteases as the propeptide is translated and processed through the default secretory pathway (Denecke et al. 1990). In some cases, the propeptide (at least the active peptide region) is modified during this process. For instance, PSKs are sulfated on the tyrosine residues in the active pentapeptide region probably as the protein is processed through the Golgi network by enzymes such as tyrosylprotein sulfotransferase (Hanai et al. 2000) before being secreted. It is likely that it is during this processing stage that proCLV3 is hydroxylated on proline residues within its active region, as a modified 12 amino acid peptide (mCLV3) containing two hydroxyproline residues derived from the CLE region of CLV3 has been identified in A. thaliana tissues (Kondo et al. 2006). The CLE motif that forms the mature hydoxyproline-mCLV3 is all that is required for its activity (Fiers et al. 2006). Similarly, C-Terminally Encoded Peptide (CEP) 1 contains hydroxyproline residues in its 14 amino acid processed form (Ohyama et al. 2008), whereas the sulfation of the tyrosine residues is required to obtain high activity of PSK-α (Matsubayashi and Sakagami 1996).
Processing and secretion has only been described for a few of the peptides and is predicted for many of the other peptides from their sequence information. However, in most cases, evidence has been obtained that at least the prototype peptide of each class is found in the apoplast (Table 1; Fig. 2), indicating that the peptide has been secreted. PSK-α was originally isolated as a cell proliferation factor in the culture medium essential for low density cell cultures of asparagus (Asparagus officinales L.) (Matsubayashi and Sakagami 1996). Other peptides have been isolated from screens for factors in the extracellular medium that stimulate defence responses such as alkalinisation of the extracellular medium; these include systemin (Pearce et al. 1991) and RALF (Pearce et al. 2001). Several of the factors were first identified from genetic screens where the knock-out mutants caused abnormal growth of particular regions such as the clv3 mutant, which contains excess stem cells in shoot apical and floral meristems that continue to enlarge over time (Clark et al. 1995). ProCLV3 is secreted into the meristematic apoplast (Rojo et al. 2002) and PNP-A is also secreted from mesophyll cells (Y. H. Wang and H. R. Irving, unpubl. obs.). EPF1 and EPF2 are involved in determining epidermal cell division events that lead to stomatal formation in leaf and stem epidermis, and were identified from mutant screens that detected abnormal stomatal patterns (Hara et al. 2007, 2009; Hunt and Gray 2009). However, an approach based on an analogy to animal systems was used to identify and purify PNP, which was immunoreactive to antisera specific for the animal peptide factor atrial natriuretic peptide (Vesely and Giordano 1991; Gehring et al. 1996; Maryani et al. 2001). The genes for PNP have since been identified (Ludidi et al. 2002) and it appears from phylogenetic data that similarities between AtPNP-A and ANP may be the result of convergent evolution (Gehring and Irving 2003).
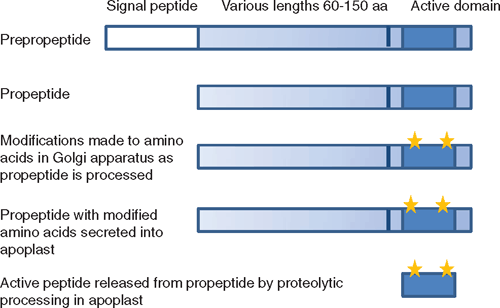
|
Once secreted into the apoplast, the propeptides can be further processed by specific extracellular proteases also secreted into the apoplast (Fig. 2). Processed peptide molecules have been identified using mass spectrometry for mCLV3, mCEP1, PSK-α and PSY1 (Matsubayashi and Sakagami 1996; Kondo et al. 2006; Amano et al. 2007; Ohyama et al. 2008; Table 1). In general, these active small peptides originate from conserved regions in the C-terminus of the propeptide molecule (Fig. 2) and this region has homology with other peptides of the same class but not between classes (Fig. 1). Alternatively, incubation of propeptide with cauliflower (Brassica oleracea L. var. botrytis L.) meristem extracts has been used to show that the propeptide is processed into active smaller peptides. This has been done with CLV3, CLE1 and Inflorescence Deficient in Abscission (IDA) to yield active mature peptide fractions (Ni and Clark 2006; Stenvik et al. 2008). Both PROPSK and PRORALF are cleaved by specific subtilisin type serine proteases in the apoplast at the dibasic amino acids upstream of the C-terminally encoded active peptide region to release the peptide (Srivastava et al. 2008, 2009). Further processing is still required to release the active pentapeptide in the case of PSK-α. Thus several enzymes and the propeptide are required to meet in the apoplast and presumably these need to be secreted from adjacent or the same cells for this to occur.
On the other hand, PNP has a region towards the N-terminus that is homologous with animal atrial natriuretic peptide and this region also contains its functional activity (Morse et al. 2004; Wang et al. 2007) but it is currently unknown if the protein is further processed. Also the low molecular weight cysteine rich (LCR) and SCRL proteins contain conserved cysteine residues throughout the secreted protein (Vanoosthuyse et al. 2001), indicating that extracellular processing may not be a universal feature of secretory peptides.
Paracrine and autocrine effects
In many cases, these signalling peptides are expressed in particular and restricted regions of the plant where they are secreted, are processed further and act upon nearby cells (Fig. 3). The receptors that have been identified are generally members of receptor-like protein families that form oligomers and recognise particular patterns of the active peptide ligand (see next two sections). So even if the peptide ligands share common receptors, their action is limited to the areas that the peptides are actually secreted and processed. This type of action is similar to growth factors regulating development in animal cells where a compound acting on adjacent or nearby cells is said to have a paracrine effect to distinguish it from that of a long distance hormone (endocrine) effect. In some cases, the compounds act on the cell that generates them, which is referred to as an autocrine effect. One of the advantages of this type of signalling is that it allows organs to respond to a gradient of molecules and is a very ancient form of signalling evident throughout the development of multi-cellular organisms. Most of the prototype peptides listed in Table 1 demonstrate paracrine and possibly autocrine signalling. This is particularly marked with those directly affecting development such as CLE, EFP and IDA, which act at specific localised regions within the plant.
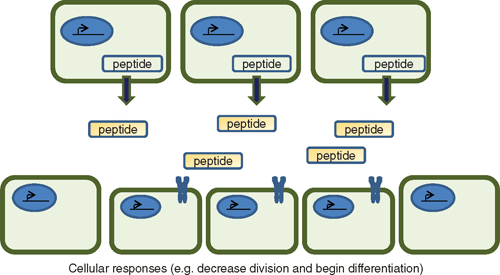
|
Peptide signalling molecules in development
Several of the peptide signalling molecules have distinct roles in development where they regulate cell differentiation and organogenesis, often by repressing cellular growth. In several instances, this has been determined from knock-out mutant studies that have identified phenotypic mutants displaying overgrowth of particular regions such as those occurring in the meristem with clv3 (Clark et al. 1995), anthers with tpd1 (Yang et al. 2003), or epidermal leaf surface and stomatal development with epf1 and epf2 mutants (Hara et al. 2007, 2009; Hunt and Gray 2009). To verify these interpretations, several groups have made use of overexpressing mutants that exhibit restricted development of the particular regions. Alternatively, peptide fractions have been directly (ectopically) applied and effects opposing those of the knock-out mutants but similar to the overexpressing mutants have been observed. For instance, overexpressing (ox) CEP1 is mainly found in the lateral root primordia and represses root growth, as does the ectopic application of synthetic mCEP1 (Ohyama et al. 2008). CLE19 is normally found in roots, and ectopic application of synthetic peptides corresponding to conserved CLE motifs of CLV3, CLE19 and CLE40 caused the termination of the root meristem, which is a similar phenotype to oxCLE19 mutants (Fiers et al. 2006). Most members of the CLE family (known as CLE-A) act to repress cell division in the meristematic regions (Whitford et al. 2008).
A useful phenomenon in genetic analysis is that similar phenotypes are associated with knock-out mutants of other members of the signalling pathway, such as the receptor and downstream signalling components. For instance, CLV3 encodes the secreted peptide that acts to repress apical and floral meristem cell division, as in its absence in the knock-out mutant clv3, the meristems continue to enlarge over time (Clark et al. 1995). While clv1 exhibits a similar and weaker phenotype (Clark et al. 1993), it encodes an LRR-RLK (Clark et al. 1997). Similarly, the receptors for IDA, EPF and TPD1 have been identified using such genetic screens and are also members of the LRR-RLK family (Hara et al. 2007; Jia et al. 2008; Stenvik et al. 2008; Hara et al. 2009). However, where genetic redundancy is evident with multigene families expressed in the same tissues, mutant screens will be considerably less useful.
Another feature that is worth remarking on is that specific peptides from a particular class involved in regulating developmental responses are expressed in quite particular and restricted regions of the plant. In addition, there is considerable redundancy in the effects of the peptide class that is, to a certain extent, counterbalanced by the restricted expression patterns. For instance, although members of the CLE-A family act to repress cell division in the meristematic regions, members of the CLE-B family (CLE41–44) affect vessel development (Whitford et al. 2008) and are homologues of the Zinnia elegans Cav. (also known as Zinnia elegans Jacq.) tracheary element differentiation factor that suppresses xylem cell differentiation in cultured mesophyll cells (Ito et al. 2006). Combinations of CLE-A and -B peptides potentiated the B-type effect of proliferation of vascular development (Whitford et al. 2008). Thus in the meristem, a reciprocal gradient of CLE-A and CLE-B-type peptides will form that will regulate organogenesis and vascular development, and may be relayed by the same or similar classes of receptors recognising different combinations of CLE ligands. This is probably not surprising, as the CLE ligand is relatively conserved (Cock and McCormack 2001); however, it is likely that multiple combinations of CLE receptors are expressed in the developing vascular and meristematic regions (also see Fukuda et al. 2007; Jun et al. 2008). Such findings highlight the importance of spatial differentiation in the expression patterns of the CLE (and other) peptides to prevent developmental errors.
Peptide signalling molecules influencing growth
Other peptides appear to have subtle effects where they may be involved in modulating general growth and development in response to the environment. Although RALF and RALFL were first identified in a screen for plant defence proteins (Pearce et al. 2001), their role is considerably more diverse and they are also likely to influence development. Exogenous application of RALFL inhibits root growth (Pearce et al. 2001) and silencing of RALF disrupts root hair development (Wu et al. 2007). In another example, PSK-α appears to act in a cooperative manner with CLE41–44. PSK-α promotes tracheary element differentiation in Z. elegans mesophyll cell cultures in the presence of auxin and cytokinin (Matsubayashi et al. 1999; Motose et al. 2009), whereas CLE41–44 inhibits this process (Ito et al. 2006; see Fukuda et al. 2007, for a discussion). PSK-α has a general proliferative effect and was discovered as a cell proliferation agent essential for low density cell cultures (Matsubayashi et al. 1996). In a further example of redundancy, in A. thaliana, there are five preproPSK genes with overlapping expression patterns throughout the plant. These proteins also seem to promote cell longevity, as plants overexpressing the PSK receptor (oxPSKR1) exhibited delayed senescence and prolonged leaf expansion; root length was reduced in pskr1 knock-out mutants (Matsubayashi et al. 2006). The effect of PSK-α on roots has recently been examined in more detail, where it was shown that PSK-α enhances root elongation by controlling cell size (Kutschmar et al. 2009). Even though the effects of PSK-α are proliferative and growth-enhancing as distinct from the growth-restricting effects of peptides affecting organogenesis, they still act in a paracrine fashion and built-in redundancy is evident.
PNP is an interesting molecule as it appears to have general effects on cellular homeostasis (Gehring and Irving 2003). PNPs represent a novel class of small proteins (~14 kDa) that are distantly related to expansions, which are regulators of cell wall extension (Ludidi et al. 2002; Kende et al. 2004). PNP also is likely to have a role in cell expansion as it enhances the volume of mesophyll protoplasts (Maryani et al. 2001; Morse et al. 2004; Wang et al. 2007). However, it appears to have many other properties as both PNP isolated from leaves and recombinant PNP-A also stimulated stomatal opening, activated the H+-ATPase and modulated ion fluxes (Pharmawati et al. 1999; Maryani et al. 2001; Ludidi et al. 2004; Wang et al. 2007), although these effects could be part of the cell expansion process. In addition, PNP protein levels are increased in NaCl-stressed whole plants and A. thaliana suspension culture cells exposed to high salt or osmoticum (Rafudeen et al. 2003). Analysis of A. thaliana microarray data through Genevestigator (Zimmermann et al. 2004) also indicates that AtPNP-A transcripts are upregulated in response to abiotic stresses such as osmoticum, salt, mineral deficiencies and ozone exposure. Recombinant PNP-A directly increases stomatal conductance and transpiration rates, which are correlated with increases in photosynthetic rates where the efficiency of light use during photosynthetic CO2 fixation was enhanced (Gottig et al. 2008). Furthermore, recombinant PNP-A modulates the effect of abscisic acid (ABA) on stomatal aperture (Wang et al. 2007). Since both compounds are upregulated in times of environmental stress (e.g. drought), it is conceivable that one of the physiological roles of PNP-A is to act as an antagonist to ABA and, in the case of stomata, promote limited gas exchange.
Peptide signalling molecules involved in defence responses
Perception of danger is a key part of the plant innate defence responses as argued by Boller and Felix (2009) in their recent review. Plants detect microbes via microbe-associated molecular signatures, which are molecules such as bacterial flagellin (flg22) with specific plant receptors that are generally members of the receptor-like proteins including members of the LRR-RLK family such as Flagellin Sensing (FLS) 2 (Boller and Felix 2009). Plants also seem to contain endogenous danger signals such as systemin, PEP1 and RALF/RALFL that are associated with responses to pathogen attack. These peptides were all identified in their active excreted processed form in alkalinisation screens (Pearce et al. 1991, 2001; Huffaker et al. 2006). Unlike systemin, both RALF and PROPEP are found throughout the plant kingdom (Pearce et al. 2001; Haruta and Constabel 2003; Germain et al. 2005; Huffaker et al. 2006; Silverstein et al. 2007). Since the processed forms of these peptides are more active, it is tempting to speculate that propeptides are found in the apoplast and the active peptides form degradation products of proteases released by damaged plant cells or the pathogens themselves. This is likely to be the case with proRALF23, which is cleaved by specific plant subtilisin serine proteases to release the active peptide (Srivastava et al. 2009). The receptor for AtPEP1 (PEPR1) has been identified through peptide crosslinking studies and it is an LRR-RLK (Yamaguchi et al. 2006) but the receptor for RALF has not yet been identified. Expression of PROPEP1 is upregulated by PEP1 itself as well as wounding, jasmonates, ethylene and bacterial flg22 (Huffaker et al. 2006), suggesting that PEP1 acts as an endogenous danger signal (Boller and Felix 2009).
PNPs may also have a role in plant defence, as coexpression and promoter content analyses indicate that PNP-A may function alongside other pathogenesis-related proteins as a component of plant defence responses (Meier et al. 2008). The functionally uncharacterised transcript from citrus CjBAp12 is similar to PNP-B and was initially isolated as a mobile peptide associated with the plant response to citrus blight, which is a disease of unknown aetiology (Ceccardi et al. 1998). Interestingly, a PNP-like gene occurs uniquely in the bacterial pathogen that causes citrus canker, Xanthomonas axonopodis pv. citri (Nembaware et al. 2004). This bacterial protein alters the plant host homeostasis responses where it increases stomatal conductance, transpiration and photosynthetic rates, and enhances the efficiency of light use during photosynthetic CO2 fixation (Gottig et al. 2008). It is speculated that expression of XacPNP allows the pathogen to create a favourable environment within the host for its growth and that it is an example of horizontal gene transfer (Gottig et al. 2008).
Receptors for peptide ligands
For ligands to communicate a message effectively, receptors need to exist that relay the message, so receptors and their ligands are thought to have evolved in parallel (Fryxell 1996). So far, the receptors for peptide ligands that have been identified are members of the receptor protein-like family and several of them are either LRR-RLKs or leucine rich repeat receptor-like proteins (LRR-RLPs). Several recent reviews have discussed the different types of receptor like proteins in relation to their interactions and signalling mechanisms (see Butenko et al. 2003; Afzal et al. 2008; Boller and Felix 2009; Tör et al. 2009). LRR-RLKs contain a large leucine rich motif that is repeated several times in the extracellular domain, a single transmembrane domain and a cytoplasmic serine-threonine kinase domain, whereas the LRR-RLPs lack the intracellular kinase domain (Fig. 4). In addition, these receptor-like proteins are sometimes associated with another group of serine-threonine kinase molecules that lack the extracellular domain (Fig. 4). Members of the LRR-RLK family that are well characterised include the brassinosteroid (AtBRI1) and flagellin (FLS2) receptors. An important part of their activation is the ability to form oligomers rapidly (within minutes) with other LRR-RLKs such as Brassinosteroid Associated Kinase 1 (BAK1) and this in turn stimulates receptor autophosphorylation and a phosphorylation cascade (Wang et al. 2005, 2008; Chinchilla et al. 2007).
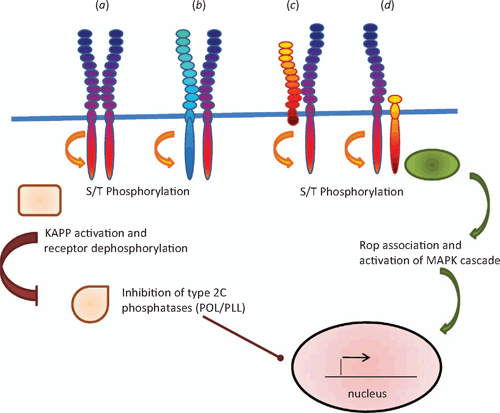
|
Oligomer formation has been identified as part of the signalling cascade in response to CLV3. CLV1 is a full length LRR-RLK (Clark et al. 1997) that directly interacts with modified CLV3 in the external leucine rich domain (Ogawa et al. 2008). CLV1 forms dimers with other closely related LRR-RLKs such as Barely Any Meristem (BAM) to bind CLV3 (DeYoung and Clark 2008) while CLV2 is receptor like molecule that lacks any kinase domain (Jeong et al. 1999) but associates with the intracellular kinase Coryne (CRN) rather than CLV1 (Müller et al. 2008; see Butenko et al. 2009 for a review). Thus two parallel receptor pathways appear to be acting in the shoot and floral meristems that are receptive to CLV3. The other peptide receptors are not so well characterised. However, receptors for PSK-α, PSY1 and PEP1 have been identified by affinity-crosslinking studies and these receptors are all LRR-RLKs (Matsubayashi et al. 2002, 2006; Yamaguchi et al. 2006; Amano et al. 2007). Mutant studies have been useful in identifying potential receptors such as the LRR-RLKs Hasea (HAS) and HAS-Like (HSL) 2 for IDA (Stenvik et al. 2008). TPD1 binds to a specific site within the extracellular leucine rich domain of Excess Microsporocytes1 (EMS1) (also known as Extra Sporogenous Cells (EXS)), and this in turn activates EMS1 receptor auto-phosphorylation (Jia et al. 2008). Similarly, Too Many Mouths (TMM), which is an LRR-RLP, and the LRR-RLKs Erecta (ER) and ER-Like (ERL) 1 and 2 were identified by mutant studies as the receptors for EPF and are thought to form an oligomer complex (Hara et al. 2007; Bhave et al. 2009; Hara et al. 2009). Several of these LRR-RLKs (PEPR1, CLV1 and ER) also contain a putative guanylate cyclase domain within the general kinase domain region (Kwezi et al. 2007) and it is of interest to speculate that production of cyclic GMP may form part of the signalling pathway in addition to the phosphorylation cascade. Indeed, in vitro studies have revealed that AtBRI1, which also contains this domain, does have guanylate cyclase activity (Kwezi et al. 2007).
Although the receptor for PNP has not been identified, the signalling cascade in response to application of PNP involves a very rapid (within seconds) increase in cGMP levels (Pharmawati et al. 1998; Pharmawati et al. 2001; Wang et al. 2007), indicating that the receptor either contains or is very closely associated with a guanylate cyclase. Rapid increases in cytoplasmic calcium occur in the surface cells of seedling roots in response to RALF1 (Haruta et al. 2008) and there is also a rapid alkalinisation of the external media, indicating that ion fluxes have been activated and ATPase-dependent proton pumps are inhibited (Pearce et al. 2001). Much remains to be discovered about the details of the signalling networks activated by the peptide signalling molecules. However, based on what is known about the BRI1–BAK interaction (Wang et al. 2005, 2008), they are likely to involve oligomerisation, phosphorylation and dephosphorylation cascades. Indeed, the CLV3–CLV1 cascade involves inhibition of the type 2C protein phosphatases Poltergeist (POL) and POL-Like (PLL), which eventually represses the transcription factor Wushel (WUS) and so inhibits stem cell formation (Mayer et al. 1998; Yu et al. 2000). A Rho-type GTPase molecule is also incorporated into the activated CLV3–CLV1 receptor complex and is thought to activate a kinase cascade (Trotochaud et al. 1999). The activity of CLV1 is reduced by a feedback system where CLV1 is dephosphorylated by the type 2C phosphatase, Kinase Associated Protein Phosphatase (KAPP) (Stone et al. 1998; Jun et al. 2008; Butenko et al. 2009). Cessation of the signal response could be further enhanced by events such as receptor mediated endocytosis, which has been reported to occur with FLS2 following 10–20 min stimulation with bacterial flagellin (Robatzek et al. 2006).
Expression of the receptors involved in organogenesis is restricted to localised areas, as occurs with CLV1, CLV2 and CRN (Müller et al. 2008). However, the receptor oligomers respond to ectopically applied ligands, which is paramount as a gradient of responses between different members of the CLE family (Whitford et al. 2008). This indicates that there is some overlapping redundancy in the receptor specificity, which is hardly surprising as they are recognising the relatively small but highly similar active peptide fragment. By the very nature of the reported actions of the ligands, it would be expected that receptors for PSK-α, PEP1, RALF and PNP are much more widely expressed. This is indeed the case for PSKR1 and 2 and PSYR1, which are widely expressed and appear to have overlapping and redundant functions, as triple mutants exhibit dwarfism due to decreases in both cell size and number (Amano et al. 2007).
Nitrogen and sulfur content of peptide signalling molecules
At first glance, it would appear counterintuitive for plants to invest heavily in nitrogen-rich molecules such as peptides and proteins for signalling molecules. Nitrogen and phosphorous are major nutrients limiting plant growth (Elser et al. 2007), and in Australian situations where ancient soils are present, phosphorous is more limiting (Lambers et al. 2009). However, a recent study revealed that plants have adapted to ecological nitrogen limitations so that crop plants have higher nitrogen contents in their transcribed RNA compared with undomesticated plants (Acquisti et al. 2009a). This phenomenon is carried through to the proteome, where crop plants and nitrogen fixing plants have proteins containing more amino acids with nitrogen-rich side chains than undomesticated plants (Acquisti et al. 2009a). Sulfur is another nutrient that, in many instances, is limiting, which will constrain the use of cysteine and methionine amino acid pools (Hawkesford and De Kok 2006).
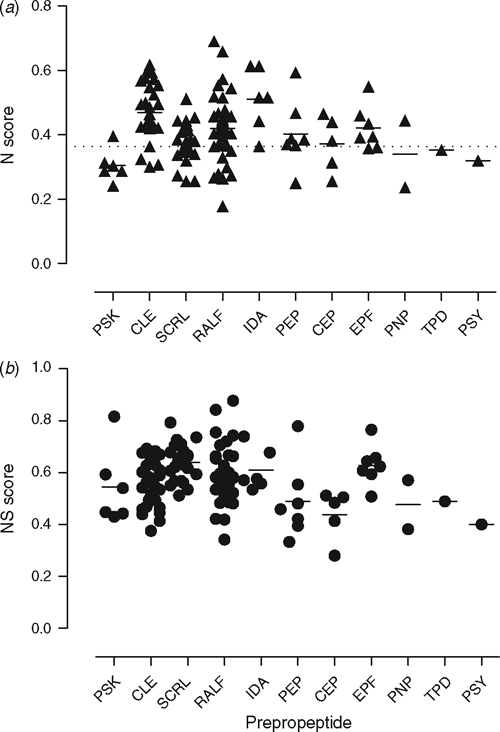
With these findings in mind, we were curious to determine if the signalling peptides (prepropeptide) were relatively nitrogen-rich, as we reasoned that their nitrogen levels may reflect their level of importance to the plant. We assessed the importance of the signalling peptides in A. thaliana (an undomesticated species) using the nitrogen proteome criteria of Acquisti et al. (2009a), who found that the average nitrogen score for all proteins in the A. thaliana proteome was 0.3637 ± 0.0788 (mean ± s.d.). Surprisingly, many of the peptides have nitrogen scores that are higher than the mean nitrogen score for the overall average of A. thaliana (Fig. 5; Table S1 (the latter is available as an Accessory Publication to this paper)). With an average of 0.413 ± 0.096 across all prepropeptides, the N value is closer to the N value reported for cellular anabolic machinery (0.482 ± 0.164) than catabolic machinery (0.313 ± 0.061) in A. thaliana (Acquisti et al. 2009b). Several of the peptides such as SCRL and RALF contain highly conserved cysteine residues (Pearce et al. 2001; Vanoosthuyse et al. 2001; Silverstein et al. 2007) and since cysteine is a structurally important amino acid and sulfur is a limiting nutrient (Hawkesford and De Kok 2006), we included sulfur containing amino acids in the analysis using a simple score based on the number of cysteine and methionine residues present, which was added to the nitrogen score to obtain a combined N–S score (Fig. 5). However, this N–S score does not include the extra processing of the propeptides such as proPSKs that are sulfated on tyrosine residues (Hanai et al. 2000). Another example of processing is the hydroxylation of proline residues in CLE and CEP (Kondo et al. 2006; Ohyama et al. 2008). It is evident that the plant peptide signalling molecules are the result of a considerable expenditure of energy and presumably represent a worthwhile investment.
|
A further factor that should be considered in this analysis is that amino acids found in proteins can be readily recycled through the cellular proteolytic machinery (e.g. proteosomes) and thus can be considered a renewable resource. In the last few years, the importance of protein degradation in regulating plant hormones has become apparent with the discovery of the auxin receptor TIR1 being the F-box subunit of the ubiquitin ligase complex SCFTIR1 (for a review, see Mockaitis and Estelle 2008). Moreover, plants can rapidly reprogramme their protein expression in response to the cellular energy status coordinated by the kinases KIN 10/11 (the A. thaliana orthologues of mammalian AMP activated kinase), where synthesis of biosynthetic genes is rapidly switched to catabolic enzymes in response to environmental cues such as dark or hypoxia (for a review, see Baena-González and Sheen 2008).
We argue that the expression of peptides as signalling molecules in localised restricted areas (e.g. the meristem) makes efficient use of limited nitrogen and sulfur resources that can be rapidly regulated in response to environmental cues by activation of gene transcription. We suggest that this process can be considered as a fine regional control mechanism that works in conjunction with classical hormones such as auxin and ABA to control growth and development. However, hormones such as ABA are not only synthesised de novo but ABA is also found as an inactive glucose ester conjugated ABA that circulates throughout the plant and that can be released by the action of β-glucosidases in the apoplast (Wasilewska et al. 2008) to produce an extremely rapid burst of the hormone in an immediate response to stressful environmental cues. In the case of peptide signalling molecules, such rapid responses are also partially cued, as propeptides are present in the apoplast. When the propeptide is digested by proteolytic enzymes, it releases the more active peptide ligand that triggers plant responses. We speculate that the propeptide is either inactively conjugated to another molecule in the apoplast or is present at such low concentrations that it does not activate signalling responses. However, upon cleavage, the mature active peptide can be recognised by its receptors at very low concentrations (e.g. CLE41/44, PEP1 and PSK-α) are active at subnanomolar concentrations (Matsubayashi and Sakagami 1996; Ito et al. 2006; Pearce et al. 2008). Peptide signalling molecules are thus dependent on the presence of additional enzymes to reach full activity, which provides a further level of control.
Conclusions and future perspectives
Peptide signalling molecules are used across all the kingdoms and form a relatively ancient evolutionary adaptation to the task of communicating between cells that has withstood the test of time and selection as they have several evolutionary advantages. First, the release of restricted amounts of material from specialised cells such as meristems to create signalling gradients can be used to regulate development. This is a feature of embryonic development that is carried through to adult multicellular organisms, which use paracrine signalling to ensure that particular regions or organs respond to the signal. Second, modular combinations of receptors can be used that allow flexibility in regulating responses to the peptide signalling molecules. Another advantage that secretion of peptide signals may offer over other molecules is that relatively rapid and controlled release can be achieved by secreting not only the propeptide but also the processing enzymes that ensure the mature peptide is released. Although the cost of synthesising these separate proteins may be relatively high, it is likely to be economical from a nitrogen and energy use perspective at least, as often only a few cells make this demand on nitrogen and energy resources.
Although much has been uncovered about the roles of peptide signalling molecules in plants over the last 20 years, there is still a great deal to discover. It is likely that more peptide molecules will be found, as many small peptides are not annotated in the databases (Silverstein et al. 2007). At this stage relatively few receptor–ligand pairs are known and it is likely that further receptors will be discovered for many of the peptide ligands (see Butenko et al. 2009). Relatively little is known about the downstream signalling pathways leading to gene expression and cellular responses, and this is an area particularly worth exploring as it will reveal fundamental insights into the control mechanisms regulating plant growth and development.
Acknowledgements
The authors thank Drs CA Gehring and DR Smyth for helpful and insightful discussions, and MA Wakeham for comments on the manuscript. Support from the Australian Research Council’s Discovery funding scheme (project numbers DP0557561 and DP0878194) is gratefully acknowledged.
Acquisti C,
Elser JJ, Kumar S
(2009a) Ecological nitrogen limitation shapes the DNA composition of plant genomes. Molecular Biology and Evolution 26, 953–956.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Acquisti C,
Kumar S, Elser JJ
(2009b) Signatures of nitrogen limitation in the elemental composition of the proteins involved in the metabolic apparatus. Proceedings of the Royal Society of London. Series B. Biological Sciences 276, 2605–2610.
| Crossref | GoogleScholarGoogle Scholar |

Afzal AJ,
Wood AJ, Lightfoot DA
(2008) Plant receptor-like serine threonine kinases: roles in signalling and plant defense. Molecular Plant-Microbe Interactions 21, 507–517.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Amano Y,
Tsubouchi H,
Shinohara H,
Ogawa M, Matsubayashi Y
(2007) Tyrosine-sulfated glycopeptide involved in cellular proliferation and expansion in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America 104, 18333–18338.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Baena-González E, Sheen J
(2008) Convergent energy and stress signaling. Trends in Plant Science 13, 474–482.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Bhave NS,
Veley KM,
Nadeau JA,
Lucas JR,
Bhave SL, Sack FD
(2009) TOO MANY MOUTHS promotes cell fate progression in stomatal development of Arabidopsis stems. Planta 229, 357–367.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Boller T, Felix G
(2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annual Review of Plant Biology 60, 379–406.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Butenko MA,
Patterson SE,
Grini PE,
Stenvik G-E,
Amundsen SS,
Mandal A, Aalen RB
(2003) INFLORESCENCE DEFICIENT IN ABSISSION controls floral organ abscission in Arabidopsis and identifies a novel family of putative ligands in plants. The Plant Cell 15, 2296–2307.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Butenko MA,
Vie AK,
Brembu T,
Aalen RB, Bones AM
(2009) Plant peptides in signalling: looking for new partners. Trends in Plant Science 14, 255–263.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Ceccardi TL,
Barthe GA, Derrick KS
(1998) A novel protein associated with citrus blight has sequence similarities to expansin. Plant Molecular Biology 38, 775–783.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Chinchilla D,
Zipfel C,
Robatzek S,
Kemmerling B,
Nurnberger T,
Jones JDG,
Felix G, Boller T
(2007) A flagellin-induced complex of the receptor FLS2 and BAK1 iniitates plant defence. Nature 448, 497–500.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Clark SE,
Running MP, Meyerowitz EM
(1993) CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development 119, 397–418.
| PubMed |

Clark SE,
Running MP, Meyerowitz EM
(1995) CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1
. Development 121, 2057–2067.

Clark SE,
Williams RW, Meyerowitz EM
(1997) The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89, 575–585.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Cock JM, McCormack S
(2001) A large family of genes that share homology with CLAVATA3. Plant Physiology 126, 939–942.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Denecke J,
Botterman J, Deblaere R
(1990) Protein secretion in plant cells can occur via a default pathway. The Plant Cell 2, 51–59.
| Crossref |
PubMed |

DeYoung BJ, Clark SE
(2008) BAM receptors regulate stem cell specification and organ development through complex interactions with CLAVATA signaling. Genetics 180, 895–904.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Dombrowski JE,
Pearce G, Ryan CA
(1999) Proteinase inhibitor-inducing activity of the prohormone prosystemin resides exclusively in the C-terminal systemin domain. Proceedings of the National Academy of Sciences of the United States of America 96, 12 947–12 952.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Elser JJ,
Bracken MES,
Cleland EE,
Gruner DS,
Harpole WS,
Hillebrand H,
Ngai JT,
Seabloom EW,
Shurin JB, Smith JE
(2007) Global analysis of nitrogen and phosphorous limitation of primary producers in freshwater, marine and terrestial ecosystems. Ecology Letters 10, 1135–1142.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Farmer EE,
Johnson RR, Ryan CA
(1992) Regulation of expression of proteinase inhibitor genes by methyl jasmonates and jasmonic acid. Plant Physiology 98, 995–1002.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Farrokhi N,
Whitelegge JP, Brusslan JA
(2008) Plant peptides and peptidomics. Plant Biotechnology Journal 6, 105–134.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Fiers M,
Golemiec E,
van der Schors R,
van der Geest L,
Li KW,
Stiekema WJ, Liu C-M
(2006) The CLAVATA3/ESR motif of CLAVATA3 is functionally independent from the nonconserved flanking regions. Plant Physiology 141, 1284–1292.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Fletcher JC,
Brand U,
Running MP,
Simon R, Meyerowitz EM
(1999) Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283, 1911–1914.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Fryxell KJ
(1996) The coevolution of gene family trees. Trends in Genetics 12, 364–369.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Fukuda H,
Hirakawa Y, Sawa S
(2007) Peptide signaling in vascular development. Current Opinion in Plant Biology 10, 477–482.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Gehring CA, Irving HR
(2003) Natriuretic peptides – a class of heterologous molecules in plants. The International Journal of Biochemistry & Cell Biology 35, 1318–1322.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Gehring CA,
Md Khalid K,
Toop T, Donald JA
(1996) Rat natriuretic peptide binds specifically to plant membranes and induces stomatal opening. Biochemical and Biophysical Research Communications 228, 739–744.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Germain H,
Chevalier E,
Caron S, Matton DP
(2005) Characterization of five RALF-like genes from Solanum chacoense provides support for a developmental role in plants. Planta 220, 447–454.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Gottig N,
Garavaglia BS,
Daurelio LD,
Valentine A,
Gehring C,
Orellano EG, Ottado J
(2008)
Xanthomonas axonopodis pv. citri uses a plant natriuretic peptide-like protein to modify host homeostasis. Proceedings of the National Academy of Sciences of the United States of America 105, 18 631–18 636.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Hanai H,
Nakayama D,
Yang H,
Matsubayashi Y,
Hirota Y, Sakagami Y
(2000) Existence of a plant tyrosylprotein sulfotransfrearase: novel plant enzyme catalyzing tyrosine O-sulfation of preprophytosulfokine in vitro. FEBS Letters 470, 97–101.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Hara K,
Kajita R,
Torii KU,
Bergmann DC, Kakimoto T
(2007) The secretory peptide gene EPF1 enforces the stomatal one-cell-spacing rule. Genes & Development 21, 1720–1725.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Hara K,
Yokoo T,
Kajita R,
Onishi T,
Yahata S,
Peterson KM,
Torii KU, Kakimoto T
(2009) Epidermal cell density is autoregulated via a secretory peptide, EPIDERMAL PATTERNING FACTOR 2 in Arabidopsis leaves. Plant & Cell Physiology 50, 1019–1031.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Haruta M, Constabel CP
(2003) Rapid alkalinization factors in poplar cell cultures. Peptide isolation, cDNA cloning and differential expression in leaves and methyl jasmonate-treated cells. Plant Physiology 131, 814–823.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Haruta M,
Monshausen G,
Gilroy S, Sussman MR
(2008) A cytoplasmic Ca2+ functional assay for identifying and purifying endogenous cell signaling peptides in Arabidopsis seedlings: identifcation of AtRALF1 peptide. Biochemistry 47, 6311–6321.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Hawkesford MJ, De Kok LJ
(2006) Managing sulphur metabolism in plants. Plant, Cell & Environment 29, 382–395.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Holton N,
Cano-Delgado A,
Harrison K,
Montoya T,
Chory J, Bishop GJ
(2007) Tomato BRASSINOSTEROID INSENSITIVE1 is required for systemin-induced root elongation in Solanum pimpinellifolium but is not essential for wound signaling. The Plant Cell 19, 1709–1717.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Huffaker A,
Pearce G, Ryan CA
(2006) An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proceedings of the National Academy of Sciences of the United States of America 103, 10 098–10 103.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Hunt L, Gray JE
(2009) The signaling peptide EPF2 controls asymmetric cell divisions during stomatal development. Current Biology 19, 864–869.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Ito Y,
Nakanoyo I,
Motose H,
Iwamoto K,
Sawa S,
Dohmae N, Fukuda H
(2006) Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science 313, 842–845.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Jeong S,
Trotochaud AE, Clark SE
(1999) The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. The Plant Cell 11, 1925–1933.
| Crossref |
PubMed |

Jia G,
Liu X,
Owen HA, Zhao D
(2008) Signaling of cell fate determination by the TPD1 small protein and EMS1 receptor kinase. Proceedings of the National Academy of Sciences of the United States of America 105, 2220–2225.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Jun JH,
Fiume E, Fletcher JC
(2008) The CLE family of plant polypeptide signaling molecules. Cellular and Molecular Life Sciences 65, 743–755.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Kende H,
Bradford KJ,
Brummell DA,
Cho HT, Cosgrove D ,
et al
.
(2004) Nomenclature for members of the expansin superfamily of genes and proteins. Plant Molecular Biology 55, 311–314.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Kondo T,
Sawa S,
Kinoshita A,
Mizuno S,
Kakimoto T,
Fukuda H, Sakagami Y
(2006) A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science 313, 845–848.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Kutschmar A,
Rzewuski G,
Stührwohldt N,
Bemmster GTS,
Inzé D, Sauter M
(2009) PSK-a promotes root growth in Arabidopsis. New Phytologist 181, 820–831.
| Crossref | GoogleScholarGoogle Scholar |

Kwezi L,
Meier S,
Mungur L,
Ruzvidzo O,
Irving H, Gehring C
(2007) The Arabidopsis thaliana brassinosteroid receptor (AtBRI1) contains a domain that functions as a guanylyl cyclase in vitro. PLoS ONE 2, e449.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Lambers H,
Mougel C,
Jaillard B, Hinsinger P
(2009) Plant-microbe-soil interactions in the rhizospehere: an evolutionary perspective. Plant and Soil 321, 83–115.
| Crossref | GoogleScholarGoogle Scholar |

Lanfermeijer FC,
Staal M,
Malinowski R,
Stratmann J, Elzenga JTM
(2008) Micro-electrode flux estimation confirms that the Solanum pimpinellifolium cu3 mutant still responds to systemin. Plant Physiology 146, 129–139.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Ludidi NN,
Heazlewood JL,
Seoighe C,
Irving HR, Gehring CA
(2002) Expansin-like molecules: novel functions derived from common domains. Journal of Molecular Evolution 54, 587–594.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Ludidi N,
Morse M,
Sayed M,
Wherrett T,
Shabala S, Gehring C
(2004) A recombinant plant natriuretic peptide causes rapid and spatially differentiated K+, Na+ and H+ flux changes in Aradidopsis thaliana roots. Plant & Cell Physiology 45, 1093–1098.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Malinowski R,
Higgins R,
Luo Y,
Piper L,
Nazir A,
Bajwa VS,
Clouse SD,
Thompson PR, Stratmann JW
(2009) The tomato’s brassinosteroid receptor BRI1 increases binding of systemin to tobacco plasma membranes, but is not involved in systemin signaling. Plant Molecular Biology 70, 603–616.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Maryani MM,
Bradley G,
Cahill DM, Gehring CA
(2001) Natriuretic peptides and immunoreactants modify osmoticum-dependent volume changes in Solanum tuberosum L. mesophyll cell protoplasts. Plant Science 161, 443–452.
| Crossref | GoogleScholarGoogle Scholar |

Matsubayashi Y, Sakagami Y
(1996) Phytosulfokine, sulfated peptides that induce the proliferation of single mesophyll cells of Asparagus officinales L. Proceedings of the National Academy of Sciences of the United States of America 93, 7623–7627.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Matsubayashi Y,
Hanai H,
Hara O, Sakagami Y
(1996) Active fragments and analogs of the plant growth factor, phytosulokine: structure-activity relationships. Biochemical and Biophysical Research Communications 225, 209–214.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Matsubayashi Y,
Takagi L,
Omura N,
Morita A, Sakagami Y
(1999) The endogenous sulfated pentapeptide phytodulfokine-a stimulates tracheary element differentiation of isolated mesophyll cells of Zinnia. Plant Physiology 120, 1043–1048.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Matsubayashi Y,
Ogawa M,
Morita A, Sakagami Y
(2002) An LRR receptor kinase involved in perception of a peptide plant hormone, phytosulfokine. Science 296, 1470–1472.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Matsubayashi Y,
Ogawa M,
Kihara H,
Niwa M, Sakagami Y
(2006) Disruption and overexpression of Arabidopsis phytosulokine receptor gene affects cellular longevity and potential for growth. Plant Physiology 142, 45–53.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Mayer FK,
Schoof H,
Haecker A,
Lenhard M,
Jurgens G, Laux T
(1998) Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95, 805–815.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

McGurl B,
Orozco-Cardenas M,
Pearce G, Ryan CA
(1994) Overexpression of the prosystemin gene in transgenic tomato plants generates a systemic signal that constitutively induces proteinase inhibitor synthesis. Proceedings of the National Academy of Sciences of the United States of America 91, 9799–9802.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Meier S,
Bastian R,
Donaldson L,
Murray S,
Bajic V, Gehring C
(2008) Co-expression and promoter content analyses assign a role in biotic and abiotic stress response to plant natriuretic peptides. BMC Plant Biology 8, 24.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Mockaitis K, Estelle M
(2008) Auxin receptors and plant development: a new signaling paradigm. Annual Review of Cell and Developmental Biology 24, 55–80.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Morse M,
Pironcheva G, Gehring C
(2004) AtPNP-A is a systemically mobile natriuretic peptide immunoanalogue with a role in Arabidopsis thaliana cell volume regulation. FEBS Letters 556, 99–103.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Motose H,
Iwamoto K,
Endo S,
Demura T,
Sakagami Y,
Matsubayashi Y,
Moore KI, Fukuda H
(2009) Involvement of phytosulfokine in the attenuation of stress response during the transdifferentiation of Zinnia mesophyll cells into tracheary elements. Plant Physiology 150, 437–447.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Müller R,
Bleckmann A, Simon R
(2008) The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. The Plant Cell 20, 934–946.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Nembaware V,
Seoighe C,
Sayed M, Gehring C
(2004) A plant natriuretic peptide-like gene in the bacterial pathogen Xanthomonas axonopodis may induce hyper-hydration in the plant host: a hypothesis of molecular mimicry. BMC Evolutionary Biology 4, 10.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Ni J, Clark SE
(2006) Evidence for functional conservation, sufficiency, and proteolytic processing of the CLAVATA3 CLE domain. Plant Physiology 140, 726–733.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Oelkers K,
Goffard N,
Weiller GF,
Gresshoff PM,
Mathesius U, Frickey T
(2008) Bioinformatic analysis of the CLE signaling peptide family. BMC Plant Biology 8, 1.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Ogawa M,
Shinohara H,
Sakagami Y, Matsubayashi Y
(2008)
Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science 319, 294.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Ohyama K,
Ogawa M, Matsubayashi Y
(2008) Identification of a biologically active, small secreted peptide in Arabidopsis by in silico gene screening, followed by LC-MS-based structure analysis. The Plant Journal 55, 152–160.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Opsahl-Ferstad HG,
Le Deunff E,
Dumas C, Rogowsky PM
(1997)
ZmESR, a novel endosperm-specific gene expressed in a restricted region around the maize embryo. The Plant Journal 12, 235–246.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Papadopoulos JS, Agarwala R
(2007) COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics 23, 1073–1079.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Pearce G,
Strydom D,
Johnson S, Ryan CA
(1991) A polypeptide from tomato leaves induces wound-inducible proteinase inhibitor proteins. Science 253, 895–897.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Pearce G,
Moura DS,
Stratmann J, Ryan CA
(2001) RALF, a 5-KDa ubiquitous polypeptide in plants, arrests growth and development. Proceedings of the National Academy of Sciences of the United States of America 98, 12 843–12 847.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Pearce G,
Yamaguchi Y,
Munske G, Ryan CA
(2008) Structure-activity studies of AtPep1, a plant peptide signal involved in the innate immune response. Peptides 29, 2083–2089.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Pharmawati M,
Gehring CA, Irving HR
(1998) An immunoaffinity purified plant natriuretic peptide analogue modulates cGMP level in the Zea mays root stele. Plant Science 137, 107–115.
| Crossref | GoogleScholarGoogle Scholar |

Pharmawati M,
Shabala SN,
Newman IA, Gehring CA
(1999) Natriuretic peptides and cGMP modulate K+, Na+ and H+ fluxes in Zea mays roots. Molecular Cell Biology Research Communications 2, 53–57.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Pharmawati M,
Maryani MM,
Nikolakopoulos T,
Gehring CA, Irving HR
(2001) Cyclic GMP modulates stomatal opening induced by natriuretic peptides and immunoreactive analogues. Plant Physiology and Biochemistry 39, 385–394.
| Crossref | GoogleScholarGoogle Scholar |

Rafudeen S,
Gxaba G,
Makgoke G,
Bradley G,
Pironcheva G,
Raitt L,
Irving H, Gehring C
(2003) A role for plant natriuretic peptide immuno-analogues in NaCl- and drought-stress responses. Physiologia Plantarum 119, 554–562.
| Crossref | GoogleScholarGoogle Scholar |

Robatzek S,
Chinchilla D, Boller T
(2006) Ligand induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes & Development 20, 537–542.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Rojo E,
Sharma VK,
Kovaleva V,
Raikhel NV, Fletcher JC
(2002) CLV3 is localized to the extracellular space, where it activates the Arabidopsis CLAVATA stem cell signaling pathway. The Plant Cell 14, 969–977.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Ryan CA, Pearce G
(1998) Systemin: a polypeptide signal for plant defensive genes. Annual Review of Cell and Developmental Biology 14, 1–17.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Scheer JM, Ryan CA
(2002) The systemin receptor SR160 from Lycopersicon peruvianum is a member of the LRR receptor kinase family. Proceedings of the National Academy of Sciences of the United States of America 99, 9585–9590.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Schilmiller AL, Howe GA
(2005) Systemic signaling in the wound response. Current Opinion in Plant Biology 8, 369–377.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Schopfer CR,
Nasrallah ME, Nasrallah JB
(1999) The male determinant of self-incompatibility in Brassica. Science 286, 1697–1700.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Silverstein KAT,
Moskal WJ,
Wu HC,
Underwood BA,
Graham MA,
Town CD, VandenBosch KA
(2007) Small cysteine-rich peptides resembling antimicrobial peptides have been under-predicted in plants. The Plant Journal 51, 262–280.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Srivastava R,
Liu J-X, Howell SH
(2008) Proteolytic processing of a precursor protein for a growth-promoting peptide by a subtilsin serine protease in Arabidopsis. The Plant Journal 56, 219–227.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Srivastava R,
Liu J-X,
Guo H,
Yin Y, Howell SH
(2009) Regulation and processing of a plant peptide hormone, AtRALF23, in Arabidopsis. The Plant Journal 59, 930–939.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Stenvik G-E,
Tandstad NM,
Guo Y,
Shi C-L,
Kristiansen W,
Holmgren A,
Clark SE,
Aalen RB, Butenko MA
(2008) The EPIP peptide of INFLORESCENCE DEFICIENT IN ABSISSION is sufficient to induce abscission in Arabidopsis through the receptor-like kinases HASEA and HASEA-LIKE2. The Plant Cell 20, 1805–1817.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Stone JM,
Trotochaud AE,
Walker JC, Clark SE
(1998) Control of meristem development by CLAVATA1 receptor kinase and kinase-associated protein phosphatase interactions. Plant Physiology 117, 1217–1225.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Tör M,
Lotze MT, Holton N
(2009) Receptor-mediated signalling in plants: molecular patterns and programmes. Journal of Experimental Botany 60, 3645–3654.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Trotochaud AE,
Hao T,
Wu G,
Yang Z, Clark SE
(1999) THe CLAVATA1 receptor-like kinase requires CLAVATA3 for its assembly into a signaling complex that includes KAPP and a Rho-l related protein. The Plant Cell 11, 393–405.
| Crossref |
PubMed |

Vanoosthuyse V,
Miege C,
Dumas C, Cock JM
(2001) Two large Arabidopsis thaliana gene families are homologous to the Brassica gene superfamily that encodes pollen coat proteins and the male component of the self-incompatibility locus. Plant Molecular Biology 46, 17–34.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Vesely DL, Giordano AT
(1991) Atrial natriuretic peptide hormonal system in plants. Biochemical and Biophysical Research Communications 179, 695–700.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Wang X,
Goshe MB,
Soderblom EJ,
Phinney BS,
Kuchar JA,
Li J,
Asami T,
Yoshida S,
Huber SC, Clouse SD
(2005) Identification and functional analysis of in vivo phosphorylation sites of the Arabidopsis BRASSINOSTEROID-INSENSITIVE1 receptor kinase. The Plant Cell 17, 1685–1703.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Wang YH,
Gehring C,
Cahill DM, Irving HR
(2007) Plant natriuretic peptide active site determination and effects on cGMP and cell volume regulation. Functional Plant Biology 34, 645–653.
| Crossref | GoogleScholarGoogle Scholar |

Wang X,
Kota U,
He K,
Blackburn K,
Li J,
Goshe MB,
Huber SC, Clouse SD
(2008) Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Developmental Cell 15, 220–235.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Wasilewska A,
Vlad F,
Sirichandra C,
Redko Y,
Jammes F,
Valon C,
Frei dit Frey N, Leung J
(2008) An update on abscisic acid signaling in plants and more… Molecular Plant 1, 198–217.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Whitford R,
Fernandez A,
De Groodt R,
Ortega E, Hilson P
(2008) Plant CLE peptides from two distinct functional classes synergistically induce division of vascular cells. Proceedings of the National Academy of Sciences of the United States of America 105, 18 625–18 630.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Wu J,
Kurten EL,
Monshausen G,
Hummel GM,
Gilroy S, Baldwin IT
(2007) NaRALF, a peptide signal essential for the regulation of root hair tip apoplastic pH in Nicotiana attenuata, is required for root hair development and plant growth in native soils. The Plant Journal 52, 877–890.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Yamaguchi Y,
Pearce G, Ryan CA
(2006) The cell surface leucine-rich repeat receptor for AtPep1, an endogenous peptide elicitor in Arabidopsis, is functional in transgenic tobacco cells. Proceedings of the National Academy of Sciences of the United States of America 103, 10 104–10 109.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Yang S-L,
Xie L-F,
Mao H-Z,
Puah CS,
Yang W-C,
Jiang L, Sundaresan V
(2003)
TAPETUM DETERMINANT1 is required for cell specialization in the Arabidopsis anther. The Plant Cell 15, 2792–2804.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Yu LP,
Simon EJ,
Trotochaud AE, Clark SE
(2000) POLTERGEIST functions to regulate meristem development downstream of the CLAVATA loci. Development 127, 1661–1670.
| PubMed |

Zimmermann P,
Hirsch-Hoffmann M,
Hennig L, Gruissem W
(2004) Genevestigator. Arabidopsis microarray database and analysis toolbox. Plant Physiology 136, 2621–2632.
| Crossref | GoogleScholarGoogle Scholar | PubMed |



