Integrated analysis of transcriptomic and proteomic data reveals novel regulators of soybean (Glycine max) hypocotyl development
Xueliang Zhang A # , Zhikang Shen B C # , Xiaohu Sun C , Min Chen
B C # , Xiaohu Sun C , Min Chen  B C * and Naichao Zhang
B C * and Naichao Zhang  A *
A *
A
B
C
Handling Editor: Gayatri Venkataraman
Abstract
Hypocotyl elongation directly affects the seedling establishment and soil-breaking after germination. In soybean (Glycine max), the molecular mechanisms regulating hypocotyl development remain largely elusive. To decipher the regulatory landscape, we conducted proteome and transcriptome analysis of soybean hypocotyl samples at different development stages. Our results showed that during hypocotyl development, many proteins were with extreme high translation efficiency (TE) and may act as regulators. These potential regulators include multiple peroxidases and cell wall reorganisation related enzymes. Peroxidases may produce ROS including H2O2. Interestingly, exogenous H2O2 application promoted hypocotyl elongation, supporting peroxidases as regulators of hypocotyl development. However, a vast variety of proteins were shown to be with dramatically changed TE during hypocotyl development, including multiple phytochromes, plasma membrane intrinsic proteins (PIPs) and aspartic proteases. Their potential roles in hypocotyl development were confirmed by that ectopic expression of GmPHYA1 and GmPIP1-6 in Arabidopsis thaliana affected hypocotyl elongation. In addition, the promoters of these potential regulatory genes contain multiple light/gibberellin/auxin responsive elements, while the expression of some members in hypocotyls was significantly regulated by light and exogenous auxin/gibberellin. Overall, our results revealed multiple novel regulatory factors of soybean hypocotyl elongation. Further research on these regulators may lead to new approvals to improve soybean hypocotyl traits.
Keywords: aquaporins, cell walls, growth regulation, hydrogen peroxide, hypocotyl development, phytochrome, proteomics, seed storage proteins, translation efficiency.
Introduction
Hypocotyl elongation is the key step of soybean seed germination. Soybean (Glycine max) seed germination is referred to as ‘epigeal’, which means during the germination process, the hypocotyl is elongated and drag the cotyledons to break the soil surface to ensure the smooth emergence of the seedling. Therefore, hypocotyl development is crucial for successful seedling establishment and subsequent plant development. The hypocotyl is also an important link between plant aerial and underground tissues, playing important roles in nutrient transport and plant adaption to the environment.
The plasticity of hypocotyl growth is achieved through a complex signalling network which are tightly regulated by genetic and environmental factors. A large number of studies conducted in Arabidopsis thaliana have revealed the mechanisms regulating hypocotyl growth at multiple levels. First, hypocotyls rapidly elongate in response to a variety of external and internal cues, such as light and phytohormones (Feng et al. 2008; Pierik et al. 2009). Light is the most important regulator of hypocotyl elongation, which is perceived and responded by plants via photoreceptors, especially phytochromes (PHYs) (Yang et al. 2009). When exposed to light, phytochrome A (PHYA), a major phytochrome perceiving far-red light, would be translocated to nucleus, where it directly interacts with suppressor of phytochrome A (SPA) to inhibit the activity of the constitutive photomorphogenic 1 (COP1)/SPA complex, thereby suppresses hypocotyl cell elongation (Sheerin et al. 2015). PHYA could also induce hypocotyl elongation through the degradation of phytochrome interacting factors (PIFs) and the production of auxin (Jia et al. 2020). Second, hypocotyl elongation is dependent on the reorganisation and extension of primary cell wall. Xyloglucan endotransglucosylase/hydrolase (XTH) is an enzyme that catalyses the cleavage and polymerisation of xyloglucan, the essential constituent of the primary cell wall. XTH was reported to participate in cell wall construction of growing tissues, including the hypocotyl (Osato et al. 2006; Miedes et al. 2013). In XTH overexpression plants, the growth of hypocotyl cells is significantly accelerated (Miedes et al. 2013). In addition, enzymes regulating modification and degradation of pectin, which composes the matrix surrounding cellulose-xyloglucan network, were also found to be involved in the regulation of hypocotyl elongation (Derbyshire et al. 2007; Pelletier et al. 2010). Third, the development of hypocotyl is affected by the metabolism of seed storage nutrients. Plants accumulate storage reserves in mature seeds, including proteins, lipids and carbohydrates, to provide essential energy and metabolic intermediates for germination and early seedling growth, including hypocotyl development. Therefore, the efficiency of storage reserve mobilisation could affect the elongation of hypocotyls. In Arabidopsis, an interesting example is that lipid mobilisation in endosperm directly affects hypocotyl elongation under dark conditions (Penfield et al. 2004). Several studies also showed that soybean seed germination and early seedling development were related to protein hydrolysis (Han et al. 2013; Delgado et al. 2015).
Like other dicots, the soybean hypocotyl derives from embryonic cells, and its elongation is also regulated by a variety of environmental factors (e.g. light, temperature), hormones (e.g. ethylene, gibberellin (GA) and auxin) and genetic mechanisms (Hatfield and Egli 1974; Jiang et al. 2020). Under field conditions, soybean hypocotyl length is determined by sowing depth, water availability and canopy shade, etc. Several genes have been reported to be involved in the regulation of soybean hypocotyl elongation. For example, in a recent study, it was shown that single knockout of several PHYAs and PHYBs through CRISPR-Cas9 would lead to longer soybean hypocotyls (Zhao et al. 2022). Another example is MYB133, which regulates the elongation of hypocotyl through PRR5 and PIF4 (Shan et al. 2021). However, the regulation mechanisms of soybean elongation remain elusive to a large extent.
Few large-scale high-throughput omics studies have been conducted to identify genes and modules involved in the regulation of soybean seed germination and hypocotyl elongation. We recently published a study that identified a PIFs-SAURs (small auxin upregulated RNAs)-expansins module regulating hypocotyl elongation using weighted gene co-expression network analysis (WGCNA) based on transcriptomic data (Shen and Chen 2022). Wang et al. (2022) clarified the regulatory genes and potential pathways by which red light regulates the growth of soybean hypocotyls using proteomic and metabolomic approaches. However, proteomic changes during soybean hypocotyl development and their relationship with transcriptome changes remain largely unexplored, which limits the in-depth understanding of the regulation of soybean hypocotyl elongation. In this study, we analysed the transcriptome and proteome of undeveloped and developing hypocotyls, identified multiple potential regulators of hypocotyl elongation, and validated the functions of some candidate regulators. Our results provide new insights into regulation of soybean post-germination development and potential targets for variety improvement.
Materials and methods
Plant material
For the study of proteome and transcriptome change during hypocotyl development, the soybean (Glycine max L.) cultivar ‘Williams 82’ was used. For the comparison of proteomes of cultivars with longer and shorter hypocotyls, cultivar ‘Liu Tiao Qing’ (named as SJZ1045 in this study), ‘Lv 75’ (SJZ190), ‘Ban Ye Sheng’ (SJZ292) and ‘PI496 A’ (SJZ1234) were used. For the ectopic expression of soybean genes in Arabidopsis thaliana L., the ecotype Col-0 was used.
All soybean seeds were germinated in vermiculite at 25°C, under dark conditions unless specified. Soybean seeds used in this study were harvested in 2018, from the same field at Shijiazhuang, Hebei Province, China, and stored at 5°C until they were germinated.
To generate transgenic lines overexpressing GmSTS, GmPHYA1 and GmPIP1-6, the full-length coding sequences (CDS) of these genes were amplified and cloned into the pCambia1300-eCFP vector using a homologous recombination-based method. Transgenic Arabidopsis lines were generated using the floral dip method. Positive transgenic lines were screened on Murashige and Skoog (MS) medium with 50 μg/mL of hygromycin, followed by real-time quantitative PCR (qPCR) for confirmation of overexpression with gene-specific primers (data not shown). The T1 transgenic seedlings were used for subsequent experiments.
Proteomics assay
Proteomes of hypocotyl samples were investigated using tandem mass tags (TMT)-based quantitative proteomic assay, performed by PTM Biolabs (Hangzhou, China). Briefly, for each sample, total protein from three biological replicates was extracted using the protamine sulfate (PS) precipitation method with trichloroacetic acid (TCA)/acetone precipitation and digested using a filter-aided sample preparation (FASP) approach. The digested peptides were collected and labelled with TMT. After fractionation and separation, the peptides were analysed with liquid chromatography tandem mass spectrometry. Data from the mass spectrometry were analysed using the MaxQuant software (ver.1.5.2.8). The relative amount of each protein was quantified using the label-free quantification (LFQ) method.
Protein extraction and quantification
For storage protein extraction, soybean hypocotyls were homogenised in liquid nitrogen with pre-cooled mortar and pestle. The homogenate was mixed with 10 times of diethyl ether and kept at room temperature for 8 h. The supernatant was then removed and the precipitate was kept and air-dried. For every 500 mg precipitate, 5 mL 0.2 M Tris-HCl buffer (pH 7.8) was added, and the mixed solution was blended at 4°C for 9 h and then centrifuged at 17 800g for 30 min. The supernatant was collected as seed storage protein solution and subjected to electrophoresis. The protein concentration was determined by the Bradford method using BSA as the standard.
Hydrogen peroxide quantification
Content of hydrogen peroxide in soybean hypocotyls was measured using a hydrogen peroxide content assay kit (Solarbio, Beijing, China) following the manufacturer’s instruction. Briefly, soybean hypocotyls were homogenised in acetone on ice and then centrifuged. The supernatant was collected, mixed with the reaction buffer and centrifuged again. The pellet was kept and dissolved in the dissolution buffer. The amount of hydrogen peroxide was calculated by measuring the absorbance at 415 nm of the solution.
Sugar extraction and quantification
Total soluble sugar, including sucrose and maltose that can be hydrolysed to reduced monosaccharides and starch that can be partially hydrolysed to glucose, was extracted and quantified using a glycogen content assay kit (Boxbio, Beijing, China) following the manufacturer’s instruction. Briefly, soybean hypocotyls were ground into powder, added with the extraction buffer and treated with boiling water bath. The suspension was centrifuged and the supernatant was collected to measure the sugar content using a standard curve method provided by the manufacturer.
Bioinformatics analysis
The R package clusterProfiler was used to perform the Gene Ontology (GO) and Kyoto Encyclopaedia of Genes and Genomes (KEGG) enrichment analysis. Fisher’s exact test was used to determine whether there are significant differences between samples. New PLACE (https://www.dna.affrc.go.jp/PLACE) was used to perform the prediction of cis-acting regulatory elements, using the 2.5 kb upstream sequence from ATG start codon of target genes.
RNA extraction and qPCR assay
Total RNA was extracted from soybean hypocotyls using a plant RNAprep Kit (TianGen, Beijing, China). RNA concentration and purity were checked with the Qubit RNA Assay Kit using Qubit 2.0 Fluorometer (Life Technologies, CA, USA). First-strand complementary DNA (cDNA) was synthesised with 0.5–1 μg total RNA using a Reverse Transcription kit (Vazyme, Nanjing, China) and used as the template for qPCR, which was performed using a CFX96 Real-Time PCR System (Bio-Rad, CA, USA). The relative expression levels of target genes were determined using the standard-curve method as described before (Shen and Chen 2022). The reference gene was a soybean ACTIN gene (GLYMA_02G091900).
Results
Translation efficiency (TE) analysis of proteins expressed in soybean hypocotyls during development
The seed germination and subsequent seedling development of soybean are accompanied by dramatic changes in transcriptome and proteome. In order to study the regulation mechanism of soybean hypocotyl development, we sampled hypocotyls from soybean dry seeds (undeveloped hypocotyls) and 48-h germinated seeds (developing hypocotyls) and sequenced the transcriptomes and proteomes. We obtained the expression data of 57 147 transcripts and 11 269 proteins, respectively (see Supplementary Fig. S1). By comparing the expression level (LFQ intensity) of each protein and its corresponding transcript abundance (fragments per kilobase of transcript per million, FPKM), we found that there was a weak positive correlation between protein and transcript abundance in undeveloped and developing hypocotyls (R value was 0.42 and 0.16, respectively) (Fig. 1a, b). This result is consistent with the earlier research (Galland and Rajjou 2015), indicating that expression of many hypocotyl proteins might be regulated by factors other than transcription, such as protein stability, protein degradation, mRNA stability and protein transport.
Protein and mRNA expression during soybean hypocotyl development. (a, b) Scatter plot of protein-vs-transcript abundance in undeveloped (a) and developing (b) hypocotyls. R denotes Pearson’s correlation coefficient. FPKM, fragments per kilobase of transcript per million; LFQ, label-free quantitation intensity. (c, d) Gene Ontology cell component (GO-CC) and Kyoto Encyclopaedia of Genes and Genomes (KEGG) analysis results of genes with extreme high TE value in undeveloped (c) and developing (d) hypocotyls.
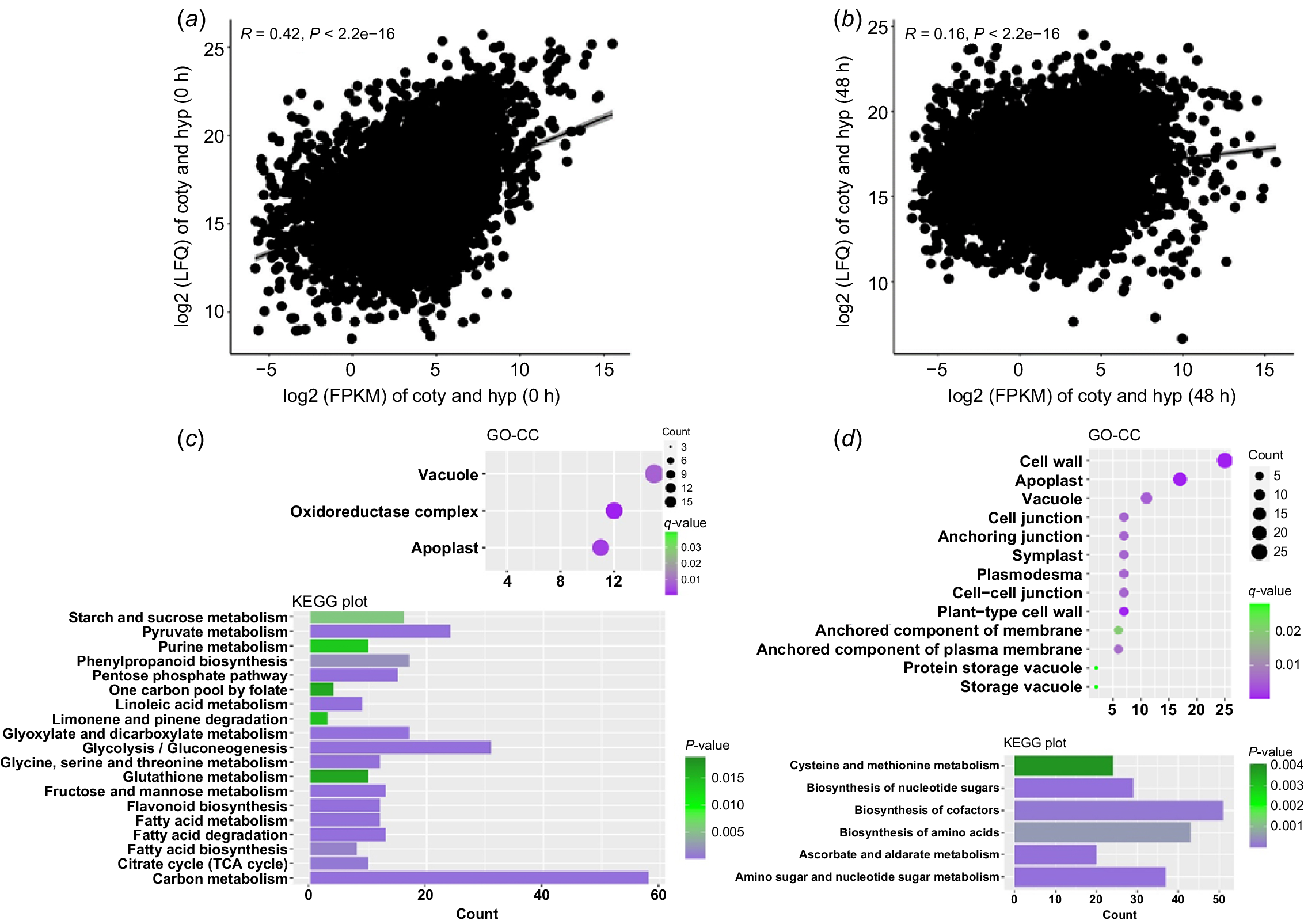
TE is the rate at which mRNA is translated into protein in cells, referred to as the rate of protein production per mRNA. Modulating TE allows the cell to fine-tune the spatiotemporal expression of genes. Several studies have shown that many proteins expressed in plant seeds are with abnormally higher or lower TE than the average level, which may play important roles in seed germination and subsequent seedling development (Galland and Rajjou 2015). By calculating the TE value of each protein in our study (TE = log2(LFQ intensity/FPKM)), we found that the overall TE is higher in developing hypocotyls (median value 12.75) than undeveloped ones (median value 11.79), indicating that proteins are more regulated by post transcription mechanisms along with hypocotyl development in general. To further study the function of proteins with abnormal TE, we selected 500 proteins with highest TE (TE > 17.5) in undeveloped and developing hypocotyls (TE > 20), respectively, where 90% of the proteins are with a TE value less than 15.6 in both situations (Fig. 1a, b).
Proteins with extreme high TE in soybean hypocotyls
The GO analysis results showed that in the undeveloped hypocotyls, most of the proteins with highest TE are storage vacuole components, including many seed storage proteins (SSPs) (Fig. 1c, Table S1). It is not surprising as SSPs are synthesised and deposited into storage vacuoles during seed maturation and stably accumulated to high levels for the preparation of germination. Consistent with this result, KEGG analysis showed that many proteins with high TE in undeveloped hypocotyls participate in the metabolism of nutrients, including amino acid, starch, sugar and fat acid (Fig. 1c). Correspondingly, in developing hypocotyls, more abnormal proteins are involved in the synthesis of amino acid, ribose and cofactors (Fig. 1d), indicating that hypocotyl development relies on the energy and metabolic intermediates derived from the mobilisation of stored nutrients, including SSPs.
Proteins with highest TE in developing hypocotyls include 10 XTHs, four pectate lyases (PELs), three pectin acetylesterases (PAEs), six pectin esterases (PEs) and four polygalacturonase (PGs) (Table S2). These enzymes were previously reported to be involved in the hydrolysis and reorganisation of cellulose molecules in cell wall; thus, allowing the expansion and growth of hypocotyl cells (Osato et al. 2006; Derbyshire et al. 2007; Pelletier et al. 2010; Miedes et al. 2013). Interestingly, by comparing the protein levels of them in soybean cultivars with different length of hypocotyls (Fig. S1, S2), we found that the XTHs were with higher protein levels in varieties with longer hypocotyls (Fig. S3a), which is consistent with their potential role in promoting hypocotyl elongation. A similar trend was observed for the PELs and PAEs, while it was opposite for the PEs (Fig. S3b), suggesting that these PAEs and PELs are positive regulators of hypocotyl elongation and PEs are negative ones.
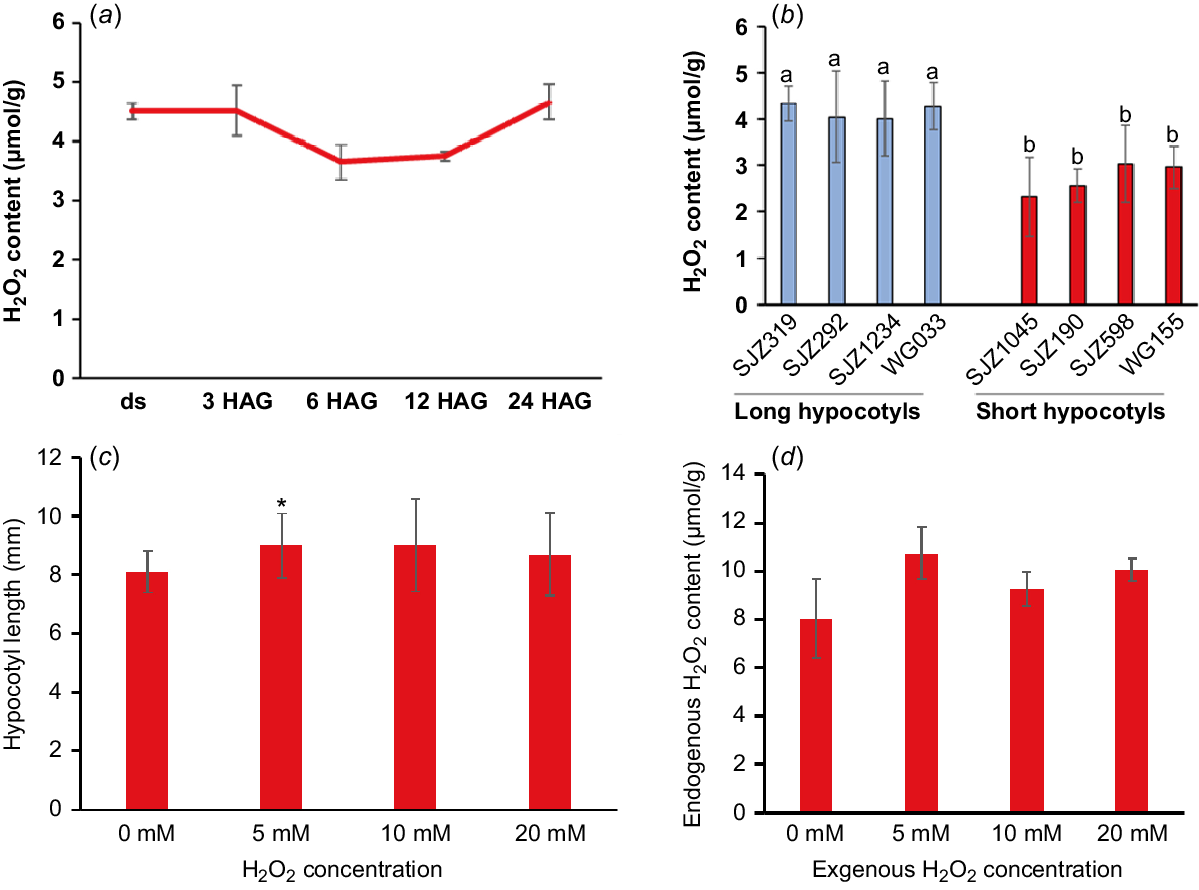
Interestingly, there were many oxidoreductases with extreme high TE in undeveloped and developing hypocotyls. For example, five lipoxygenases and four superoxide dismutases (SODs) were found to be with highest TE in undeveloped hypocotyls (Table S1) and 34 peroxidases with similar pattern were found in developing hypocotyls (Table S2). These enzymes could catalyse the formation of reactive oxygen species (ROS), including hydrogen peroxide (H2O2), or use H2O2 as the electron acceptor to catalyse downstream oxidative reactions (Quan et al. 2008; Petrov and Van Breusegem 2012). Previously reports have shown that intracellular H2O2 can break off polysaccharides to loosen the cell wall structure and play a vital role in the cell elongation growth (Schopfer 1996). Other results suggested that H2O2 could selectively oxidises proteins and mRNAs during post-germination seedling establishment (Wojtyla et al. 2016). These results lead us to hypothesise that H2O2 play a role in promoting soybean hypocotyl elongation. In our study, H2O2 was shown to be maintained a high level during the development of soybean hypocotyls (Fig. 2a) and the overall H2O2 content and peroxidase protein levels is higher in varieties with longer hypocotyls (Figs 2b and S3c). Most importantly, exogenous application of H2O2 significantly promoted the hypocotyl elongation while increasing the endogenous H2O2 content (Fig. 2c, d). All these results showed that H2O2 is indeed a positive regulator of soybean hypocotyl elongation and the enzymes related to its production and metabolism may play important roles in hypocotyl development.
H2O2 is a promoting factor of soybean hypocotyl elongation. (a) Endogenous H2O2 concentration (μmol/g) in soybean hypocotyls dissected from dry seeds (ds) and seedlings germinated for 3, 6, 9, 12 and 24 h (hours after germination, HAG). (b) H2O2 concentration in 5-day-old hypocotyls of different varieties (with longer and shorter hypocotyls, respectively). (c) Exogenous application of H2O2 promoted the elongation of hypocotyls. The hypocotyl length was measured 3 days after germination on water agar (0.9%) medium without or with different levels of H2O2. (d) Exogenous H2O2 increased the endogenous H2O2 content in 3-day-old hypocotyls. Data are presented as mean ± s.d. with n = 3 in (a), n ≥ 5 in (b) and n ≥ 10 in (c). Different lower case letters indicate significant difference in (b). *P < 0.05 relative to mock (without treatment) in (c).
Proteins with dramatic TE change during hypocotyl development
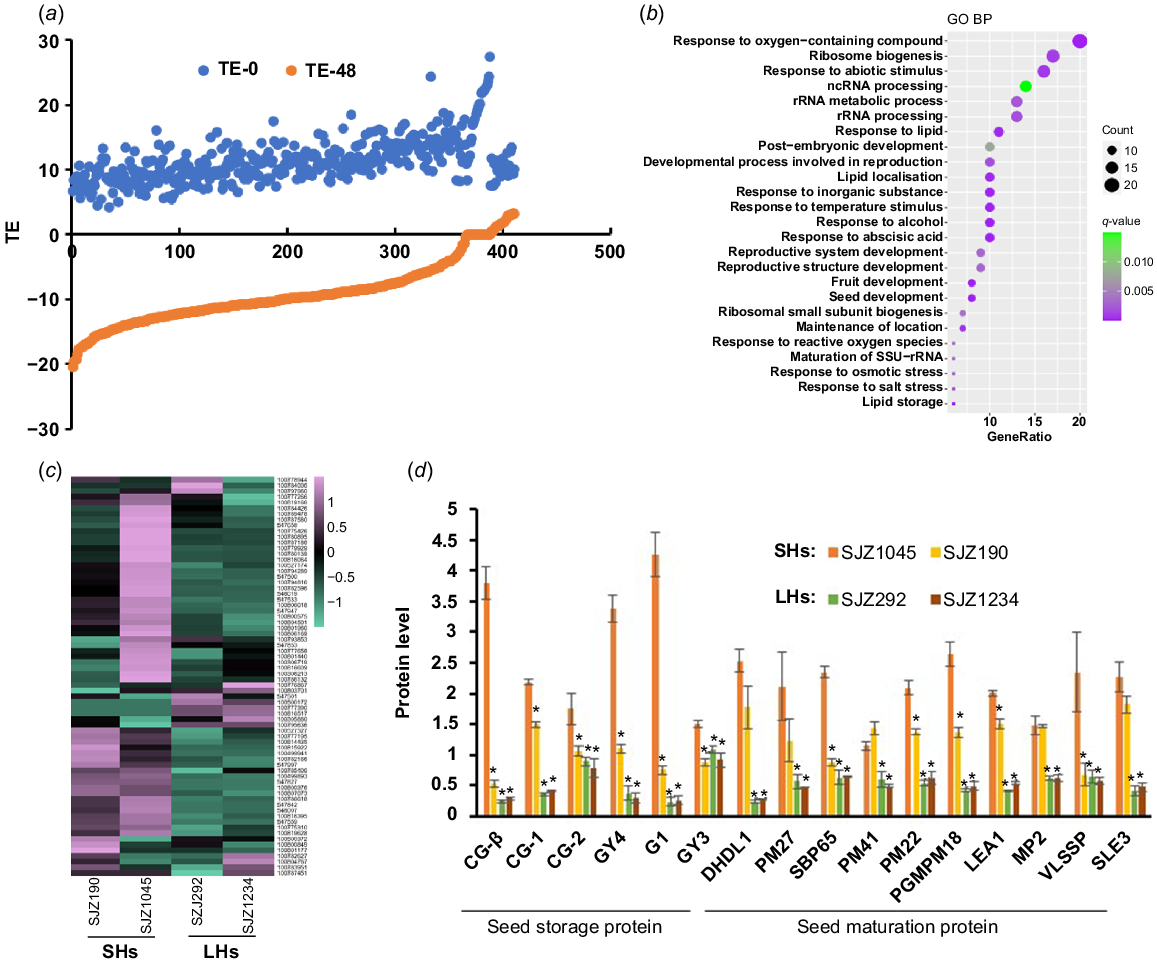
Many proteins were shown to be with dramatically changed TE during hypocotyl elongation in our experiments (Figs 3a and 4a), which may be associated with soybean hypocotyl development. We therefore selected the proteins with significant increase (TE48-TE0 > 5) (Fig. 3a) and decrease (TE48-TE0 < −5) (Fig. 4a) of TE for further study, where the TE value of 95% of the proteins changed between −3.3 and 3.3. It is worth noting that during the development of hypocotyls, the number of proteins with increased TE value was much bigger than those with decreased TE, reflecting the exuberant protein synthesis activity during hypocotyl development.
Proteins with rapidly decreased TE during hypocotyl development. (a) A large number of proteins in hypocotyls are with rapidly decreased TE along with germination. (b) GO analysis result of these proteins. (c) Expression heat map of these proteins in 4-day-old hypocotyls of different varieties. (d) Expression of selected proteins in 4-day-old hypocotyls of different varieties. Data are from label-free quantitative proteomic data and presented as mean ± s.d. (n = 3). *P < 0.05 relative to SJZ1045. The protein/gene names corresponding to the abbreviations are in Table S3. SHs, short hypocotyls; LHs, long hypocotyls.
Proteins with rapidly increased TE during hypocotyl development. (a) A large number of proteins in hypocotyls are with rapidly increased TE along with germination. (b) GO analysis result of these proteins. (c) Expression heat map of these proteins in 4-day-old hypocotyls of different varieties. (d) Expression of selected proteins in 4-day-old hypocotyls of different varieties. Data are from label-free quantitative proteomic data and presented as mean ± s.d. (n = 3). *P < 0.05 relative to SJZ1045. The protein/gene names corresponding to the abbreviations are in Table S4. SHs, short hypocotyls; LHs, long hypocotyls.
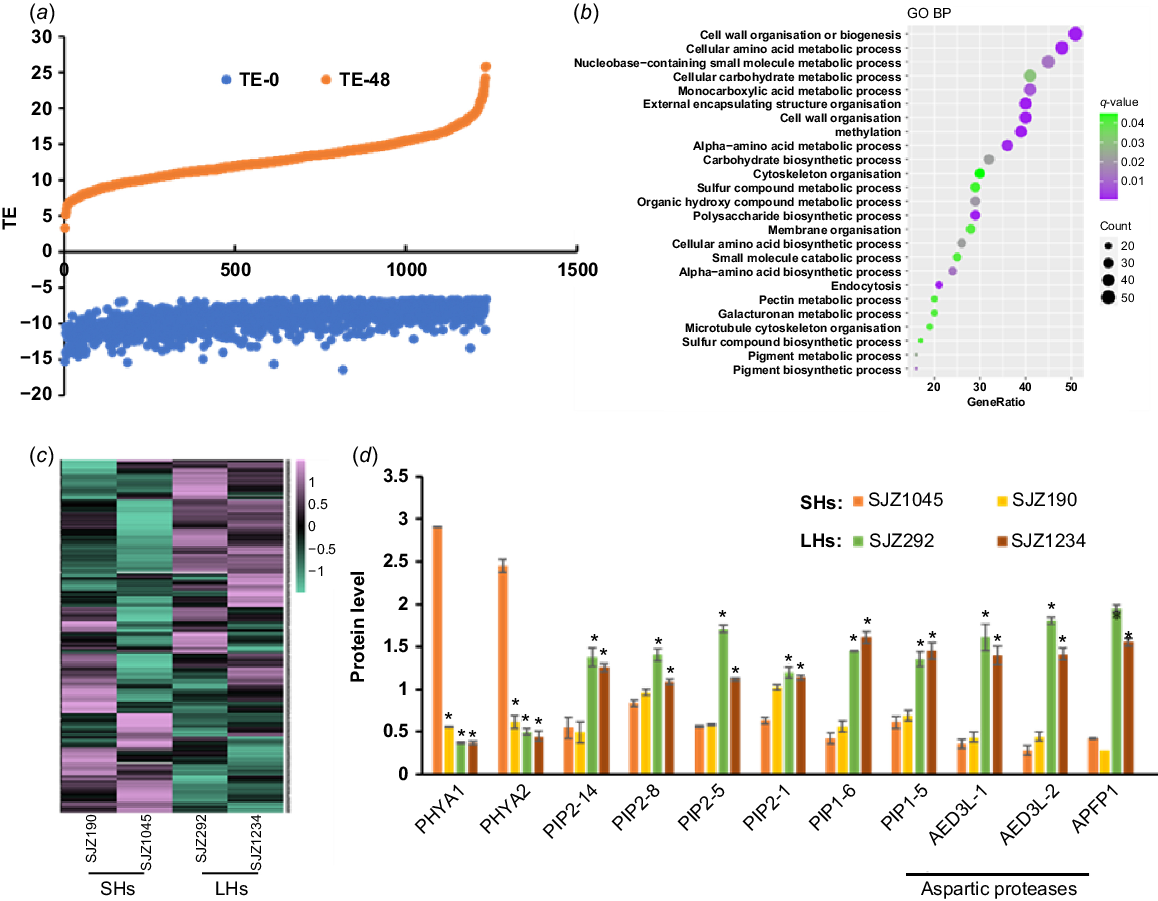
Proteins with rapidly decreased TE during hypocotyl development
Proteins with decreased TE are more likely to play a role during the early stage of seed germination (imbibition and nutrition digestion). The decrease of TE is usually due to protein degradation and/or translation stagnation. In fact, the abundances of proteins with significant decreased TE were shown to descend rapidly during hypocotyl development (Fig. S4a), while the corresponding transcript levels went down synchronously (Fig. S4b), indicating that these proteins are more likely to be degraded along with hypocotyl development. GO analysis showed that these proteins are mainly involved in response to abiotic stimulus and ribosome biogenesis/RNA binding (Fig. 3b), implying that they may be accumulated in undeveloped hypocotyl for the stress response and the preparation for translation initiation upon germination. Besides, many seed maturation proteins (late embryogenesis abundant (LEA) proteins, dehydrins, etc.) (Table S3) were found to be included in this category, which were reported to be involved in responses to stress such as desiccation during seed development and early germination (Battaglia and Covarrubias 2013). Interestingly, most of these seed maturation proteins were with higher expression in cultivars with shorter hypocotyls (Fig. 3c, d). Previous studies have shown that dehydrins and LEA proteins play important roles in plant response to abiotic stresses (Battaglia and Covarrubias 2013; Sun et al. 2021). Thus, the increased expression of these proteins in shorter hypocotyl samples may reflect a higher level of stress response, which may consume additional energy and inhibit hypocotyl elongation.
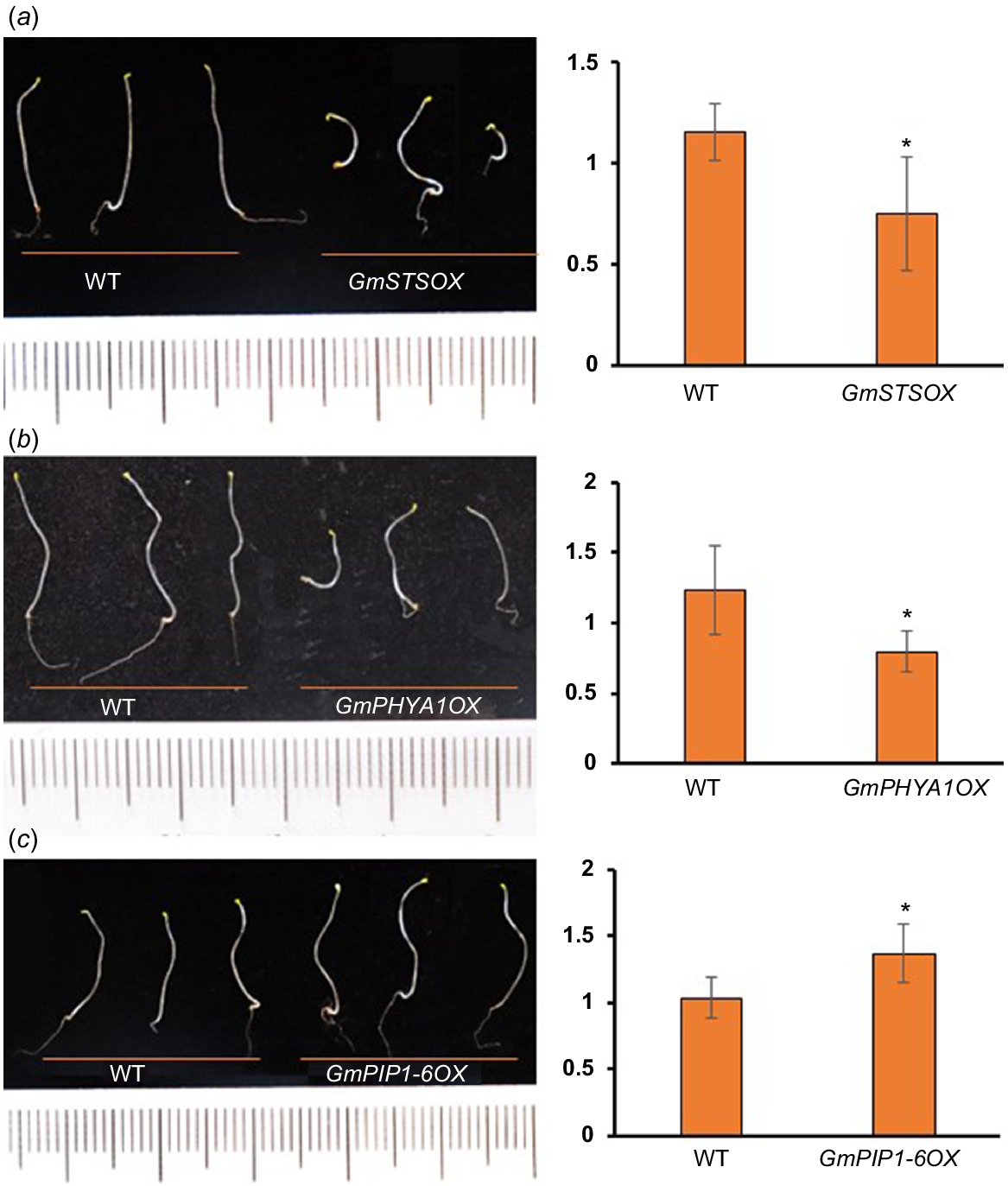
Proteins with reduced TE also include a lot of SSPs, which were thought to be accumulated during embryogenesis and rapidly mobilised upon seed germination to fuel organ development (Golombek et al. 2001; Müntz et al. 2001). As mentioned above, many of SSPs are with extreme high TE in undeveloped hypocotyls (Table S1). It is not surprising that abundances of these proteins were decreased rapidly upon seed germination, as well their transcripts, which partly explains the decrease of TE levels (Fig. S4). Strikingly, the abundances of residual SSPs were much higher in developing hypocotyls of cultivars with longer hypocotyls than shorter ones, indicating that the mobilisation efficiency of SSPs is positively related to the hypocotyl elongation (Fig. 3d). Consistent with this result, many enzymes related to protein degradation were shown to be with significant increased TE during hypocotyl development (see below), while a potential negative regulator of storage nutrient metabolism (GLYMA_19G217700, encoding a stachyose synthase), was shown to be with significant decreased TE (Table S3). Ectopic expression of this soybean stachyose synthase (STS) in Arabidopsis led to shorter hypocotyls (Fig. 5a), confirming that the mobilisation efficiency of seed storage substances regulates hypocotyl elongation.
Proteins with elevated TE during hypocotyl development
Most proteins with dramatically increased TE were shown to be with increased protein abundances along with hypocotyl development (Fig. 4a), while the corresponding transcript levels did not change as much (data not shown), indicating that the increase of TE levels may be attributed to increased protein productivity and/or less protein degradation. GO analysis showed that many of these proteins are involved in cell wall organisation and sugar/amino acid metabolic processes (Fig. 4b), which may contribute to the rapid growth at the late stage of germination. The rapidly increased production rate of these proteins ensured the fast hypocotyl growth upon germination.
Proteins with increased TE showed diverse differences between varieties with longer and shorter hypocotyls (Fig. 4c). Some were shown to be expressed higher in varieties with shorter hypocotyls, including UDP glycosyltransferase (UGTs) encoded by GLYMA_19G187400 and GLYMA_08G244700, and multiple stress or disease responsive proteins, such as thaumatin-like proteins encoded by GLYMA_08G053600 and GLYMA_05G245800, a stress response protein encoded by GLYMA_13G318600, a malate synthase encoded by GLYMA_17G128000, a glycine-rich protein encoded by GLYMA_13G244800 and an immune related protein encoded by GLYMA_19G176700 (Fig. 4c, Table S4). Previous studies have shown that UGTs play key roles in modulating auxin dependent hypocotyl elongation via buffering auxin homeostasis in Arabidopsis (Mateo-Bonmatí et al. 2021); therefore, the phenotype of shorter hypocotyls may be related to the excessive expression of these UGTs. For stress or disease resistance proteins, as hypocotyls are vulnerable structures compared to seeds, their increased expressions may be required to protect hypocotyls from adverse environment and pathogens. However, considering that the stress and disease resistance processes are energy consuming and might compete with growth and development, it is easy to understand that these proteins are expressed more highly in shorter hypocotyl varieties.
Other proteins were expressed higher in cultivars with shorter hypocotyls, including three aspartic proteases, two plasma membrane intrinsic proteins (PIPs) and two PHYAs (Fig. 4c, d, Table S4). It is worth noting that for these aspartic proteases, PIPs, PHYs and their homologues, most of them were with elevated TE during hypocotyl development (for aspartic proteases, 17 out of 22; for PIPs, 7 out of 8; for phytochromes, 3 out of 3), indicating that these proteins may play important roles in hypocotyl elongation. Among them, aspartic proteases are involved in the degradation of SSPs and participate in regulating seed germination as reported in Arabidopsis (Shen et al. 2018). In our results, we found that the protein and transcript abundances of these aspartic proteases were quickly increased during hypocotyl development, especially for two of them, GLYMA_06G093600 and GLYMA_18G090900. Our data showed that their transcription and translation were both started over from zero upon seed germination and may be responsible for the mobilisation of SSPs during hypocotyl development. Consistent with this assumption, the cultivars with longer hypocotyls contained more carbohydrates as the product of protein mobilisation in the developing hypocotyls (Fig. S5b), although there is no significant difference of SSPs in undeveloped hypocotyls from all the varieties (Fig. S5a). Thus, the protein mobilisation efficiency in the cultivars with longer hypocotyls may be more sufficient to promote hypocotyl elongation.
For the two PHYA homologues (PHYA1 and PHYA2) with increased TE (Fig. S6a), it was shown that their protein and mRNA abundances were increased in the wake of hypocotyl development (Fig. S6b), suggesting that they were actively transcribed and translated upon seed germination. Our results showed that these two soybean PHYAs were with higher protein abundances in the cultivars with shorter hypocotyls (Fig. 4c, d, Table S4), indicating their inhibitory role in hypocotyl development, which is consistent with the results in Arabidopsis (Yang et al. 2009; Sheerin et al. 2015). Not surprisingly, ectopic expression of PHYA1 in Arabidopsis caused shorter hypocotyls (Fig. 5b). Therefore, these two PHYAs are negative regulators of hypocotyl elongation.
For the two PIPs (PIP2-14 and PIP1-6) with rapidly elevated TE (Fig. S6), they were with higher expression in the varieties with longer hypocotyls (Fig. 4c, d, Table S4). PIPs belong to plant major intrinsic protein (MIP) family and act as water channels through plasma membrane, playing important roles in plant water homeostasis. Plant PIPs were reported to be involved in multiple biological processes, including nutrient transport, seed germination and the lateral root emergence (Péret et al. 2012; Liu et al. 2013; Lu et al. 2018). There are 22 genes encoding PIPs in soybean genome, including eight PIP1 and 14 PIP2. Interestingly, there are four other PIPs (PIP2-8, PIP2-5, PIP2-1 and PIP1-5) with higher expression in the cultivars with longer hypocotyls (Fig. 4c, d, Table S4), indicating that PIPs may participate in soybean hypocotyl development by regulating water transport during cell elongation. To confirm this, we overexpressed PIP1-6 in Arabidopsis, and found that the hypocotyls of transgenic plant seedlings were significantly longer than those of wild-type (Fig. 5c). Therefore, these soybean PIPs may play as positive regulators of hypocotyl elongation.
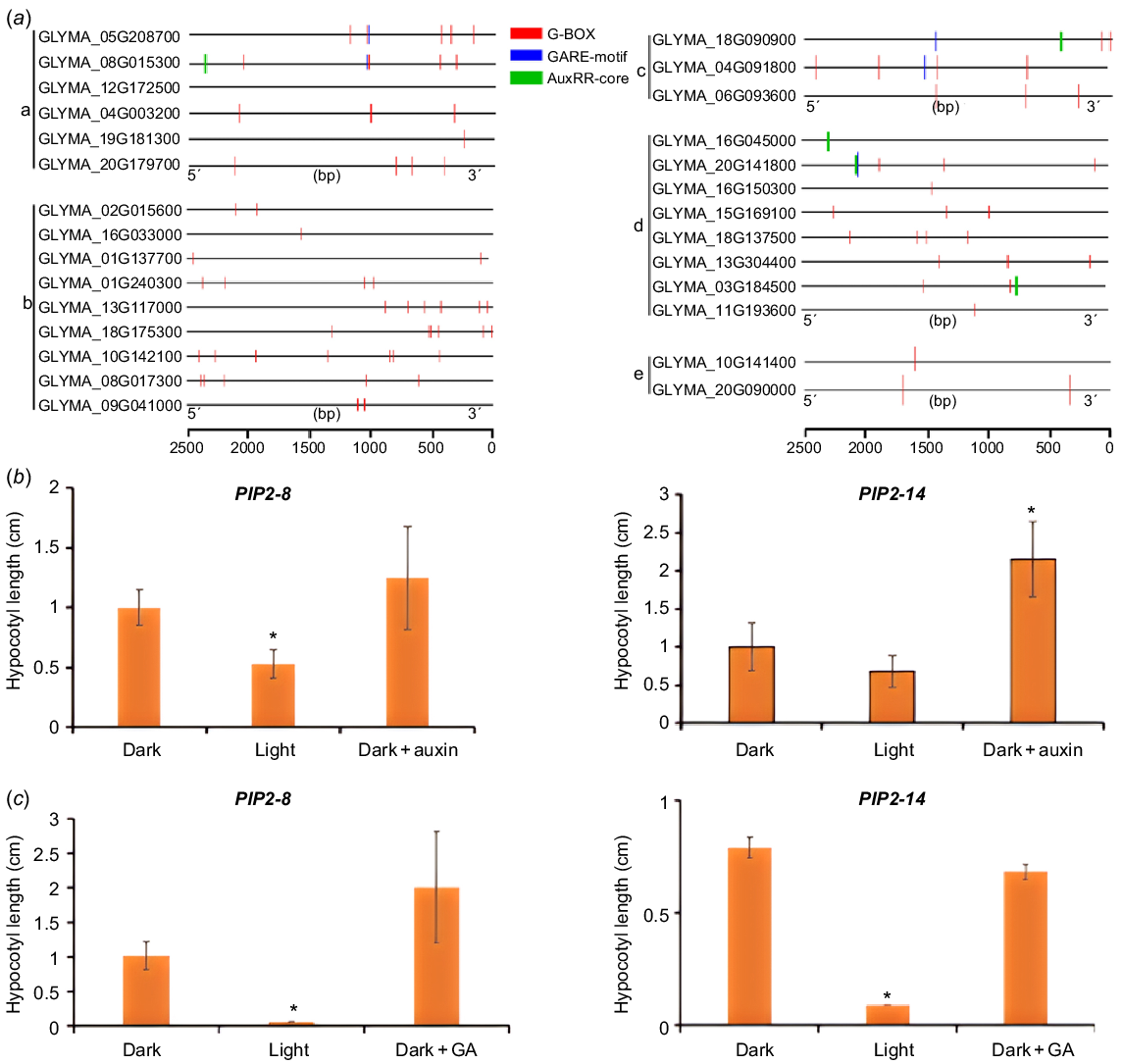
The regulation of gene expression is mostly carried out through transcription factors binding to cis elements on promoters. By analysing the upstream promoters of genes encoding the above potential regulators of hypocotyl development, we found that most of promoters contain at least one of the elements responsive to light (e.g. G-box, a specific binding site for PIF transcription factors) and phytohormones (e.g. GARE motif, a cis acting element in response to GA; AuxRR core, a binding site for ARFs) (Fig. 6a and S7), suggesting that environmental factors and hormone signals may regulate hypocotyl elongation through these regulators (Fig. 6a). To confirm this, we measured the expression of two PIPs in hypocotyls with light and auxin/GA treatment. We found that light could indeed inhibit the expression of PIP2-8 and PIP2-14 in hypocotyls (Fig. 6b, c), and exogenous auxin could induce the expression of these two genes (Fig. 6b), but the exogenous GA didn’t show significant effect (Fig. 6c).
The potential regulators of hypocotyl elongation are under regulation of light and phytohormones. (a) The promoter regions (2.5 kb) of these potential regulators contain multiple light, auxin and GA responsive elements. a: PIPs, b: PGs, c: aspartic proteases, d: XTHs, e: PHYAs. (b) Expression of two GmPIPs in the hypocotyl after 1-day treatment of light or 2 h of exogenous 10 μM IAA. (c) Expression of two GmPIPs in the hypocotyl after 1-day treatment of light or 2 h of exogenous 5 μM GA3. *P < 0.05 relative to the negative control (without treatment).
Discussion
Hypocotyl elongation is the key step of soybean seedling establishment upon seed germination. By analysing the proteome and transcriptome changes during the development of soybean hypocotyl, we identified multiple novel regulators of hypocotyl development, including XTHs, H2O2, aspartic proteases, PIPs and PHYAs. These potential regulators may participate in cell wall loosening, redox signalling, SSP metabolism, water transport and light perception, respectively. Some of them have been reported in other species, while others were rarely studied, but their functions in soybean seed germination and organ development are still not quite clear. Our research revealed their important roles in the development of soybean hypocotyl for the first time.
Soybean seeds synthesise and store a large amount of SSPs to prepare for germination, which were accumulated at very high protein but low corresponding mRNA levels (Han et al. 2013; Delgado et al. 2015). Along with seed germination and subsequent hypocotyl development, SSPs are rapidly degraded and utilised to provide precursors and energy for post germination growth; thus, the mobilisation of SSPs upon seed gemination may play a vital role in early seedling development. Our data confirmed that SSPs were of extreme high TE values in dry seeds (Fig. 3a). Upon germination, SSPs were quickly mobilised as SSP mobilisation related enzymes were with rapidly increased TE (Fig. 4a) and a negative regulator of SSP mobilisation showed the potential to suppress the hypocotyl elongation (Fig. 5a). These results support that SSP mobilisation may control the hypocotyl elongation. Interestingly, multiple SSP mobilisation enzyme encoding genes were predicted to contain multiple light and hormone responsive elements in their promoters (Fig. 6a), indicating that SSP mobilisation may be affected by external and internal factors to regulate hypocotyl elongation.
Sugar serves as not only direct energy and carbon source, but also an essential regulator for hypocotyl growth. In Arabidopsis, sugar was reported to regulates auxin, GA and BR signalling during hypocotyl elongation, through the PIF mediated gene regulation or TOR (target of rapamycin) dependent energy and nutrient responses (Xue et al. 2021). In soybean, our data implied the role of sugar as a direct energy source in hypocotyl development (Fig. S6b). Previously, sugar was reported to promote the formation of cellulose as the key component of cell wall, but there are few reports on sugar signalling during growth and development. Lu et al. (2014) reported a gene involved in sucrose signalling, GmRAV, affect hypocotyl development in the presence of exogenous sucrose. A study by Wang et al. (2023) suggests that blue light could promote the synthesis of carbohydrates and inhibit the hypocotyl elongation. Therefore, in soybean, sugar may act as both nutrient source and signalling molecule in regulating hypocotyl development. Further studies should help clarify the hidden link between sugar signalling and soybean hypocotyl elongation.
Cell wall expansion is the physical basis of hypocotyl elongation. XTHs and pectin degradation/methylation related enzymes (PELs, PAEs, PEs and PGs) control the loosening and reorganisation of cell wall by regulating the connection between cellular microfibrils in cell wall. Previous studies have shown that XTHs are involved in regulating the elongation of hypocotyls and internodes in Arabidopsis and rice (Oryza sativa) (Osato et al. 2006; Miedes et al. 2013). Our results showed that XTHs are with extreme high TE in developing soybean hypocotyls while expressed much higher in varieties with longer hypocotyls (Table S2, Fig. S3a), consistent with the previous finding that the inhibition of soybean hypocotyl elongation was associated with decreased XTH activities under low water potential (Wu et al. 2005). Thus, XTHs may act as promoters of soybean hypocotyl elongation. In addition, in rice and Arabidopsis, XTHs have been reported to be regulated by GA and contain GA responsive elements in their promoters (Jan et al. 2004; Cao et al. 2006). In soybean, we found that at least three XTHs contain GARE elements in their promoters, while all the promoters contain light responsive G-box elements (Fig. 6a), indicating that they act downstream of light and GA to regulate hypocotyl elongation, similar to their homologues in Arabidopsis and rice. In addition, pectin degradation/methylation related enzymes including PELs and PAEs were shown to be with similar expression patterns to XTHs (Table S2, Fig. S3b) and all contain G-box elements in the promoters of their encoding genes (Fig. 6a), suggesting that they may be as well positive regulators of soybean hypocotyl elongation.
H2O2 has been reported to control organ development via modifying cell wall structure or acting as a redox signal by selectively oxidising target proteins (Schopfer 1996; Huang et al. 2011; Wojtyla et al. 2016). Our results showed that multiple enzymes related to the production of H2O2 were with extreme high TE during hypocotyl development (Table S1) and expressed more in varieties with longer hypocotyls (Fig. S3c), indicating the relationship between H2O2 production and soybean hypocotyl elongation. The result that exogenous H2O2 application promoted the elongation of soybean hypocotyls (Fig. 3c) confirmed H2O2 as a positive regulator of soybean hypocotyl development.
PHYAs are photoreceptors that perceive and transduce light signals to regulate plant growth, including hypocotyl development. Our results showed that soybean PHYAs may act as negative regulators of hypocotyl elongation (Figs S6a, 4d and 5b). A recent study has shown that nuclear localised PHYA could induce constitutive photomorphogenesis phenotype i.e. shorter hypocotyl (Li and Hiltbrunner 2021). Other studies showed that in Arabidopsis and rice, the expression of PHYA is repressed by light and PHYA in turn regulates GA signalling and hypocotyl elongation. In soybean, the regulatory mechanisms may be similar, which would be confirmed by further investigation.
PIPs are known for their capacity to control cell expansion during plant elongation growth by regulating water transport. Our results showed that at least two soybean PIPs (PIP2-14 and PIP1-6) are potential promoters of hypocotyl elongation, as they were highly expressed during hypocotyl development (Fig. S6a) and the expression levels were much higher in varieties with longer hypocotyls (Fig. 4d). Their regulatory roles were confirmed by that ectopic expression of PIP1-6 in Arabidopsis promoted hypocotyl elongation (Fig. 5c). Interestingly, a study in yeast has shown that water-permeable aquaporins are also H2O2 channels (Almasalmeh et al. 2014), and a recent study showed that sorghum PIP2;1 can transport H2O2 to regulate plant response to alkaline stress (Zhang et al. 2023). Therefore, it is possible that soybean PIPs transduce putative oxidative signal during hypocotyl development, which could be investigated in future research. Besides, overexpression and/or knockout of these PIPs in soybean, as well as the previously mentioned PHYAs and STS, will help us better understand the functions and related regulatory mechanisms of these genes.
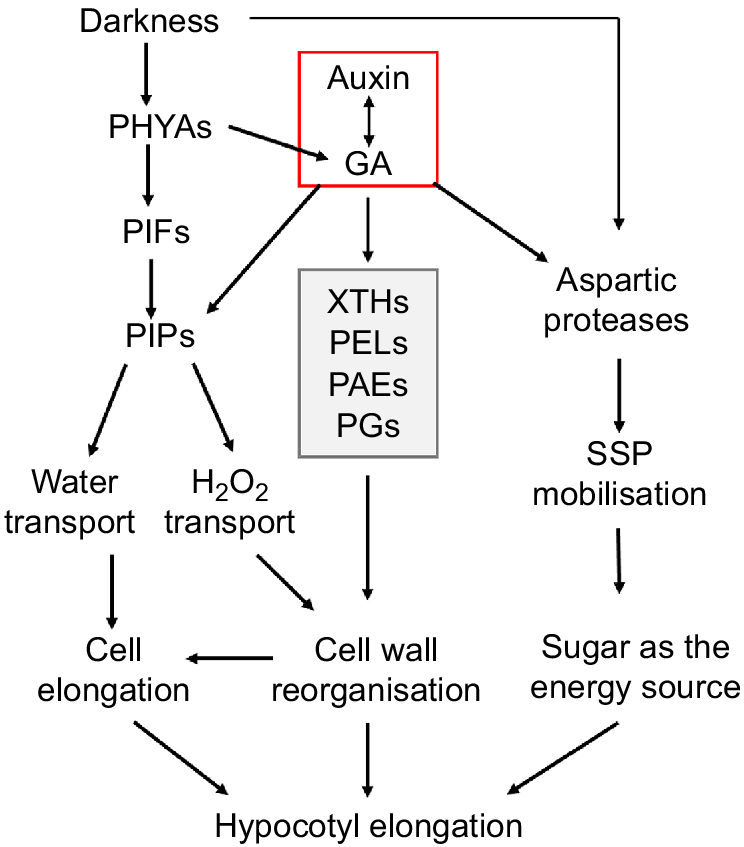
Gibberellin and auxin are key plant hormones that promote hypocotyl elongation under shade conditions. Bawa et al. (2020) showed that weak light significantly induced the biosynthesis of gibberellin and auxin, thus promoting the elongation of soybean hypocotyls. In this process, gibberellin and auxin influenced each other’s synthesis mutually. In this study, we found that most of the potential regulatory genes contain at least one light/GA/auxin responsive element in their promoters (Fig. 6a), and the expression of two PIP genes (as examples) were really regulated by light and exogenous auxin (Fig. 6b). Therefore, auxin may play a central role in regulating soybean hypocotyl elongation under low light, through PIP dependent water/H2O2 transport, cellulose hydrolysis dependent cell wall loosening and protease dependent SSP mobilisation. However, the effect of GA cannot be excluded (Fig. 7). This is consistent with our previous result that auxin regulates soybean hypocotyl elongation through GmPIFs/GmPRE6s and the downstream EXPANSIN genes (Shen and Chen 2022).
A schematic diagram showing the regulatory network underpinning soybean hypocotyl elongation. External environmental cues (darkness) and hormones (auxin, GA) can promote the water transport dependent on PIPs and/or cell wall cellulose hydrolysis and reorganisation relying on key enzymes such as XTHs, thus promoting the loosening and elongation of cell wall. Darkness and hormones may also promote the elongation of the hypocotyl by increasing storage protein mobilisation through proteolytic enzymes.
Hypocotyl development is a complex process regulated by multiple environmental factors and internal genetic mechanisms. Our research has demonstrated novel potential regulatory factors of soybean hypocotyl development at multiple levels for the first time and preliminarily established a comprehensive regulatory network including hormone and light signal perception, redox signal transduction, water transport, SSP mobilisation and cell wall reorganisation. Further analysis of relevant regulatory mechanisms in the future will reveal more detailed mechanisms of soybean hypocotyl development, providing novel targets for soybean variety improvement.
Data availability
The original contributions presented in the study are publicly available. This transcriptomics data can be found in National Center for Biotechnology Information (NCBI) BioProject database under accession number SRR17239198. The proteomics data presented in the study could be found in ProteomeXchange database under accession number PXD036677.
Declaration of funding
This research was funded by National Natural Science Foundation of China (grant number 31970344) and Joint Funds of the Natural Science Foundation of Hainan Province (grant number 2021JJLH0065).
References
Almasalmeh A, Krenc D, Wu B, et al. (2014) Structural determinants of the hydrogen peroxide permeability of aquaporins. The FEBS Journal 281(3), 647-656.
| Crossref | Google Scholar | PubMed |
Battaglia M, Covarrubias AA (2013) Late Embryogenesis Abundant (LEA) proteins in legumes. Frontiers in Plant Science 4, 190.
| Crossref | Google Scholar |
Bawa G, Feng L, Chen G, et al. (2020) Gibberellins and auxin regulate soybean hypocotyl elongation under low light and high-temperature interaction. Physiologia Plantarum 170(3), 345-356.
| Crossref | Google Scholar | PubMed |
Cao D, Cheng H, Wu W, et al. (2006) Gibberellin mobilizes distinct DELLA-dependent transcriptomes to regulate seed germination and floral development in Arabidopsis. Plant Physiology 142(2), 509-525.
| Crossref | Google Scholar | PubMed |
Delgado CML, Coelho CMMd, Buba GP, et al. (2015) Mobilization of reserves and vigor of soybean seeds under desiccation with glufosinate ammonium. Journal of Seed Science 37, 154-161.
| Crossref | Google Scholar |
Derbyshire P, McCann MC, Roberts K (2007) Restricted cell elongation in Arabidopsis hypocotyls is associated with a reduced average pectin esterification level. BMC Plant Biology 7, 31.
| Crossref | Google Scholar | PubMed |
Feng S, Martinez C, Gusmaroli G, et al. (2008) Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451, 475-479.
| Crossref | Google Scholar | PubMed |
Galland M, Rajjou L (2015) Regulation of mRNA translation controls seed germination and is critical for seedling vigor. Frontiers in Plant Science 6, 284.
| Crossref | Google Scholar | PubMed |
Golombek S, Rolletschek H, Wobus U, et al. (2001) Control of storage protein accumulation during legume seed development. Journal of Plant Physiology 158, 457-464.
| Crossref | Google Scholar |
Han C, Yin X, He D, et al. (2013) Analysis of proteome profile in germinating soybean seed, and its comparison with rice showing the styles of reserves mobilization in different crops. PLoS ONE 8, e56947.
| Crossref | Google Scholar | PubMed |
Hatfield JL, Egli DB (1974) Effect of temperature on the rate of soybean hypocotyl elongation and field emergence. Crop Science 14, 423-426.
| Crossref | Google Scholar |
Huang A-X, She X-P, Cao B-H, et al. (2011) Distribution of hydrogen peroxide during adventitious roots initiation and development in mung bean hypocotyls cuttings. Plant Growth Regulation 64, 109-118.
| Crossref | Google Scholar |
Jan A, Yang G, Nakamura H, et al. (2004) Characterization of a xyloglucan endotransglucosylase gene that is up-regulated by gibberellin in rice. Plant Physiology 136(3), 3670-3681.
| Crossref | Google Scholar | PubMed |
Jia Y, Kong X, Hu K, et al. (2020) PIFs coordinate shade avoidance by inhibiting auxin repressor ARF18 and metabolic regulator QQS. New Phytologist 228(2), 609-621.
| Crossref | Google Scholar |
Jiang H, Shui Z, Xu L, et al. (2020) Gibberellins modulate shade-induced soybean hypocotyl elongation downstream of the mutual promotion of auxin and brassinosteroids. Plant Physiology and Biochemistry 150, 209-221.
| Crossref | Google Scholar | PubMed |
Li J, Hiltbrunner A (2021) Is the Pr form of phytochrome biologically active in the nucleus? Molecular Plant 14(4), 535-537.
| Crossref | Google Scholar | PubMed |
Liu C, Fukumoto T, Matsumoto T, et al. (2013) Aquaporin OsPIP1;1 promotes rice salt resistance and seed germination. Plant Physiology and Biochemistry 63, 151-158.
| Crossref | Google Scholar | PubMed |
Lu Q, Zhao L, Li D, et al. (2014) A GmRAV ortholog is involved in photoperiod and sucrose control of flowering time in soybean. PLoS ONE 9(2), e89145.
| Crossref | Google Scholar | PubMed |
Lu L, Dong C, Liu R, et al. (2018) Roles of soybean plasma membrane intrinsic protein GmPIP2;9 in drought tolerance and seed development. Frontiers in Plant Science 9, 530.
| Crossref | Google Scholar |
Mateo-Bonmatí E, Casanova-Sáez R, Šimura J, et al. (2021) Broadening the roles of UDP-glycosyltransferases in auxin homeostasis and plant development. New Phytologist 232(2), 642-654.
| Crossref | Google Scholar | PubMed |
Miedes E, Suslov D, Vandenbussche F, et al. (2013) Xyloglucan endotransglucosylase/hydrolase (XTH) overexpression affects growth and cell wall mechanics in etiolated Arabidopsis hypocotyls. Journal of Experimental Botany 64(8), 2481-2497.
| Crossref | Google Scholar | PubMed |
Müntz K, Belozersky MA, Dunaevsky YE, et al. (2001) Stored proteinases and the initiation of storage protein mobilization in seeds during germination and seedling growth. Journal of Experimental Botany 52, 1741-1752.
| Crossref | Google Scholar | PubMed |
Osato Y, Yokoyama R, Nishitani K, et al. (2006) A principal role for AtXTH18 in Arabidopsis thaliana root growth: a functional analysis using RNAi plants. Journal of Plant Research 119, 153-162.
| Crossref | Google Scholar | PubMed |
Pelletier S, Van Orden J, Wolf S, et al. (2010) A role for pectin de-methylesterification in a developmentally regulated growth acceleration in dark-grown Arabidopsis hypocotyls. New Phytologist 188, 726-739.
| Crossref | Google Scholar | PubMed |
Penfield S, Rylott EL, Gilday AD, et al. (2004) Reserve mobilization in the Arabidopsis endosperm fuels hypocotyl elongation in the dark, is independent of abscisic acid, and requires PHOSPHOENOLPYRUVATE CARBOXYKINASE1. The Plant Cell 16, 2705-2718.
| Crossref | Google Scholar | PubMed |
Péret B, Li G, Zhao J, et al. (2012) Auxin regulates aquaporin function to facilitate lateral root emergence. Nature Cell Biology 14, 991-998.
| Crossref | Google Scholar | PubMed |
Petrov VD, Van Breusegem F (2012) Hydrogen peroxide – a central hub for information flow in plant cells. AoB Plants 2012, pls014.
| Crossref | Google Scholar |
Pierik R, Djakovic-Petrovic T, Keuskamp DH, et al. (2009) Auxin and ethylene regulate elongation responses to neighbor proximity signals independent of gibberellin and DELLA proteins in Arabidopsis. Plant Physiology 149, 1701-1712.
| Crossref | Google Scholar | PubMed |
Quan L-J, Zhang B, Shi W-W, et al. (2008) Hydrogen peroxide in plants: a versatile molecule of the reactive oxygen species network. Journal of Integrative Plant Biology 50(1), 2-18.
| Crossref | Google Scholar | PubMed |
Schopfer P (1996) Hydrogen peroxide-mediated cell-wall stiffening in vitro in maize coleoptiles. Planta 199, 43-49.
| Crossref | Google Scholar |
Shan B, Wang W, Cao J, et al. (2021) Soybean GmMYB133 inhibits hypocotyl elongation and confers salt tolerance in Arabidopsis. Frontiers in Plant Science 12, 764074.
| Crossref | Google Scholar | PubMed |
Sheerin DJ, Menon C, zur Oven-Krockhaus S, et al. (2015) Light-activated phytochrome A and B interact with members of the SPA family to promote photomorphogenesis in Arabidopsis by reorganizing the COP1/SPA complex. The Plant Cell 27, 189-201.
| Crossref | Google Scholar | PubMed |
Shen Z, Chen M (2022) Deciphering novel transcriptional regulators of soybean hypocotyl elongation based on gene co-expression network analysis. Frontiers in Plant Science 13, 837130.
| Crossref | Google Scholar | PubMed |
Shen W, Yao X, Ye T, et al. (2018) Arabidopsis aspartic protease ASPG1 affects seed dormancy, seed longevity and seed germination. Plant and Cell Physiology 59(7), 1415-1431.
| Crossref | Google Scholar | PubMed |
Sun Z, Li S, Chen W, et al. (2021) Plant dehydrins: expression, regulatory networks, and protective roles in plants challenged by abiotic stress. International Journal of Molecular Sciences 22(23), 12619.
| Crossref | Google Scholar | PubMed |
Wang C, Qiu H, Chen Y, et al. (2022) Red light regulates metabolic pathways of soybean hypocotyl elongation and thickening. Environmental and Experimental Botany 199, 104890.
| Crossref | Google Scholar |
Wang C, Chen Y, Cui C, et al. (2023) Blue light regulates cell wall structure and carbohydrate metabolism of soybean hypocotyl. International Journal of Molecular Sciences 24, 1017.
| Crossref | Google Scholar | PubMed |
Wojtyla Ł, Lechowska K, Kubala S, et al. (2016) Different modes of hydrogen peroxide action during seed germination. Frontiers in Plant Science 7, 66.
| Crossref | Google Scholar | PubMed |
Wu Y, Jeong B-R, Fry SC, et al. (2005) Change in XET activities, cell wall extensibility and hypocotyl elongation of soybean seedlings at low water potential. Planta 220, 593-601.
| Crossref | Google Scholar | PubMed |
Xue X, Yu Y-C, Wu Y, et al. (2021) Locally restricted glucose availability in the embryonic hypocotyl determines seed germination under abscisic acid treatment. New Phytologist 231, 1832-1844.
| Crossref | Google Scholar | PubMed |
Yang SW, Jang I-C, Henriques R, et al. (2009) FAR-RED ELONGATED HYPOCOTYL1 and FHY1-LIKE associate with the Arabidopsis transcription factors LAF1 and HFR1 to transmit phytochrome A signals for inhibition of hypocotyl elongation. The Plant Cell 21, 1341-1359.
| Crossref | Google Scholar | PubMed |
Zhang H, Yu F, Xie P, et al. (2023) A Gγ protein regulates alkaline sensitivity in crops. Science 379(6638), eade8416.
| Crossref | Google Scholar |
Zhao F, Lyu X, Ji R, et al. (2022) CRISPR/Cas9-engineered mutation to identify the roles of phytochromes in regulating photomorphogenesis and flowering time in soybean. The Crop Journal 10, 1654-1664.
| Crossref | Google Scholar |