Alternative oxidase: an inter-kingdom perspective on the function and regulation of this broadly distributed ‘cyanide-resistant’ terminal oxidase
Allison E. McDonaldDepartment of Biology, The University of Western Ontario, Biological and Geological Sciences Building, London, Ontario N6A 5B7, Canada. Email: amcdon27@uwo.ca
Functional Plant Biology 35(7) 535-552 https://doi.org/10.1071/FP08025
Submitted: 9 February 2008 Accepted: 11 July 2008 Published: 21 August 2008
Abstract
Alternative oxidase (AOX) is a terminal quinol oxidase located in the respiratory electron transport chain that catalyses the oxidation of quinol and the reduction of oxygen to water. However, unlike the cytochrome c oxidase respiratory pathway, the AOX pathway moves fewer protons across the inner mitochondrial membrane to generate a proton motive force that can be used to synthesise ATP. The energy passed to AOX is dissipated as heat. This appears to be very wasteful from an energetic perspective and it is likely that AOX fulfils some physiological function(s) that makes up for its apparent energetic shortcomings. An examination of the known taxonomic distribution of AOX and the specific organisms in which AOX has been studied has been used to explore themes pertaining to AOX function and regulation. A comparative approach was used to examine AOX function as it relates to the biochemical function of the enzyme as a quinol oxidase and associated topics, such as enzyme structure, catalysis and transcriptional expression and post-translational regulation. Hypotheses that have been put forward about the physiological function(s) of AOX were explored in light of some recent discoveries made with regard to species that contain AOX. Fruitful areas of research for the AOX community in the future have been highlighted.
Additional keywords: electron transport chain, endosymbiosis, mitochondrial respiration, reactive oxygen species, stress response, sulfide.
What is alternative oxidase?
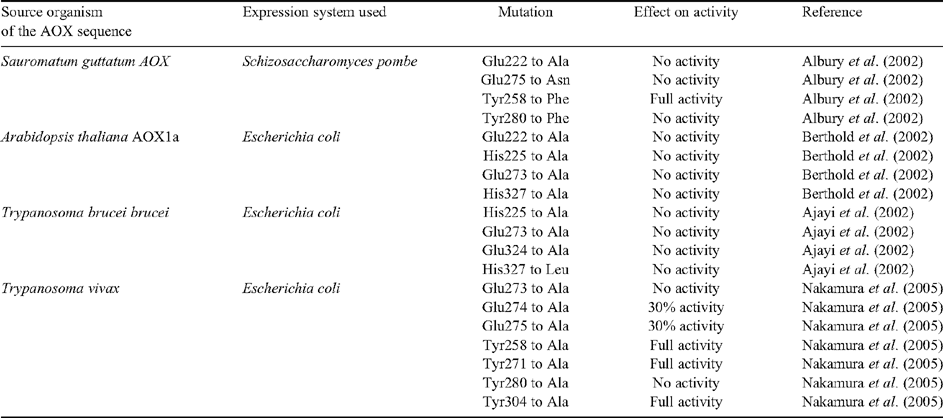
The alternative oxidase (AOX) pathway was first identified in plants and some other organisms as mitochondrial respiration (characterised by oxygen consumption) that was resistant to cyanide (CN), in contrast to the pathway utilising cytochrome c oxidase (COX), which is CN sensitive (Henry and Nyns 1975). The AOX protein is a terminal quinol oxidase in the respiratory chain of some organisms that is non-proton pumping, reduces oxygen to water, and dissipates the change in free energy as heat (Berthold et al. 2000; Berthold and Stenmark 2003; Fig. 1). AOX is resistant to inhibition by nitric oxide, azide, and sulfide, all potent inhibitors (like CN) of COX (Azcón-Bieto et al. 1989; Huang et al. 2002; Fig. 1). The compounds salicylhydroxamic acid (SHAM) and n-propyl gallate (nPG) have been shown to be effective AOX inhibitors (Vanlerberghe et al. 1994; Yip and Vanlerberghe 2001; Fig. 1).
|
Taxonomic distribution of AOX
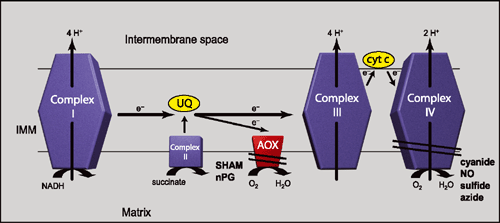
Prior to 2000, AOX had been identified in several species of plants, fungi and some protists. A major paradigm shift occurred in 2003 with the discovery of the first prokaryotic AOX (Stenmark and Nordlund 2003). Recently, AOX distribution has been demonstrated to be extremely widespread and AOX is found in all kingdoms of life except the Archaebacteria (McDonald and Vanlerberghe 2006; Fig. 2).
|
AOX genes have now been identified in many proteobacteria (Stenmark and Nordlund 2003; McDonald and Vanlerberghe 2005, 2006; Fig. 2). An analysis of a whole-genome, shotgun-sequencing metagenomic dataset derived from marine microbes in the Sargasso Sea (Venter et al. 2004) found 69 different AOX genes (most of which are believed to be prokaryotic), which approximately doubled the number of these genes in public databases at the time (McDonald and Vanlerberghe 2005). The further discovery of other eubacterial AOX sequences has demonstrated that, to date, proteobacteria are the only bacterial group known to possess AOX because it has not been seen in bacterial groups such as Actinobacteria, Chloroflexi or Firmicutes (McDonald and Vanlerberghe 2006). Results obtained from various metagenome datasets indicate that environmental sampling will continue to be an excellent source for the recovery of new and diverse sequences and this is likely to extend to AOX (Schloss and Hendelsman 2005; Tringe and Rubin 2005).
AOX is present in members of many eukaryotic lineages lacking plastids. AOX is found in several jakobids (Fig. 2), including Reclinomonas americana, a free-living mitochondriate protist whose mitochondrial genome contains more genes than any other eukaryote, suggesting that jakobids represent one of the earliest diverging eukaryotic lineages (Archibald et al. 2002). AOX is also found in the Lobosea and Heterlobosea (e.g. Dictyostelium discoideum, Hyperamoeba dachnaya and Hartmanella vermiformis) and is widespread in the fungal groups Ascomycota and Basidiomycota (McDonald and Vanlerberghe 2006; Fig. 2). This fungal distribution can now be further extended to the more basal Zygomycota and Chytrids (McDonald and Vanlerberghe 2006; Fig. 2).
AOX is present in two Ichthyosporeans, Capsaspora owczarzaki and Sphaeroforma arctica, as well as in the choanoflagellate Monosiga ovata (McDonald and Vanlerberghe 2006; Fig. 2). Molecular phylogenies have demonstrated that ichthyosporeans diverged after fungi, but before choanoflagellates and animals, and that the choanoflagellates are considered to be the sister group of animals (Lang et al. 2002).
Until recently it was assumed that AOX was not present in the Animalia (McDonald and Vanlerberghe 2004). However, AOX genes have been found in 20 animal species, representing eight different phyla; Chordata, Hemichordata, Echinodermata, Nematoda, Mollusca, Annelida, Cnidaria and Porifera (McDonald and Vanlerberghe 2006; Fig. 2). The identification of putative AOXs in several sponges (e.g. Ephydatia muelleri, Oscarella carmela and Reniera sp.), members of the most basal animal phylum, suggests that the presence of AOX is the ancestral state, and that AOX was subsequently lost by vertebrates and arthropods (McDonald and Vanlerberghe 2006). A more complete story of animal AOX evolution and distribution should emerge once more genomes become publicly available.
The three primary plastid lineages, glaucocystophyta, the red algal lineage and the green algal lineage, all possess members that contain AOX (McDonald and Vanlerberghe 2006; Fig. 2). Several cases exist where organisms that gained red or green plastids via secondary and tertiary endosymbiotic events contain AOX, and such organisms include diatoms, trypanosomes, oomycetes and dinoflagellates (McDonald and Vanlerberghe 2006; A. E. McDonald, unpubl. data; Fig. 2). AOX is widespread in the plant kingdom. Basal groups such as bryophytes and tracheophyta contain members that have AOX, as do gymnosperms (McDonald and Vanlerberghe 2006; Fig. 2). Both monocot and dicot angiosperms contain AOX and it is often present as a multigene family in individual species (McDonald and Vanlerberghe 2006; Fig. 2).
Despite the fact that AOX is widespread in nature, there are examples of members within some of these taxa that lack AOX. For example, within the Trypanosomatidae, Trypanosoma brucei brucei and Phytomonas sp. have AOX, but Leishmania sp. does not (Van Hellemond et al. 1998; McDonald and Vanlerberghe 2006). The green alga Chlamydomonas reinhardtii has AOX, but its close relative Polytomella sp. does not (Reyes-Prieto et al. 2002). Although AOX is widespread in yeast, it is absent in Saccharomyces cerevisiae and Schizosaccharomyces pombe (Veiga et al. 2003). The nematode Meliodogyne hapla has AOX, but a homology search of available nematode genomes and EST databases indicates that AOX is not present in Caenorhabditis elegans or several other nematodes (McDonald and Vanlerberghe 2006). No AOX sequence can be recovered in the subphylum vertebrata, despite the fact that several vertebrate genomes have been completed and a large number of ESTs exist. Degenerate animal AOX primers were also unsuccessful in recovering AOX in the basal vertebrates lamprey and hagfish or in several arthropod groups (A. E. McDonald and G. C. Vanlerberghe, unpubl. data). This indicates that lineage-specific losses of AOX have occurred and careful comparison of the metabolism and physiology of the organisms that lack AOX might represent another means of determining the physiological role of AOX.
Current endosymbiotic theory proposes that the mitochondrion arose from a proteobacterial ancestor and, therefore, several groups have been suggested as an endosymbiotic origin for AOX in the eukaryotic lineage (Finnegan et al. 2003; McDonald et al. 2003; Atteia et al. 2004). An analysis of the taxonomic distribution of AOX supports this model and argues for a vertical mode of inheritance of AOX in eukaryotes as opposed to multiple horizontal gene transfer events (McDonald and Vanlerberghe 2006; Fig. 2). Based on its broad taxonomic distribution, it is likely that the function(s) of AOX is important enough to have withstood selective pressure over evolutionary time and that this has led to the retention of AOX in a large number of the eukaryotic lineages that exist today.
How does AOX work?
Enzymatic function: AOX is a quinol terminal oxidase
AOXs from diverse organisms have been heterologously expressed in model bacterial or fungal systems, revealing that they function as quinol oxidases. Expression of AOX in a heme-deficient strain of Escherichia coli allows mitochondrial respiration to occur in the absence of heme-containing terminal oxidases (e.g. COX). The AOXs of the bacterium Novosphingobium aromaticivorans, the protist Cryptosporidium parvum, the protist Trypanosoma brucei brucei and the fungus Aspergillus fumigatus have all been demonstrated to be active quinol terminal oxidases when expressed in this E. coli system (Nihei et al. 2003; Stenmark and Nordlund 2003; Suzuki et al. 2004; Magnani et al. 2007). Two yeast species that lack AOX have also been used as heterologous expression systems. The AOX of the plant Sauromatum guttatum Schott (voodoo lily) is able to confer CN-resistant respiration in isolated mitochondria in the yeast Schizosaccharomyces pombe and the AOXs of the fungi Pichia (Hansenula) anomala and Aspergillus fumigatus are functional when expressed in S. cerevisiae (Crichton et al. 2005; Mathy et al. 2006; Magnani et al. 2007). The AOX of the tunicate animal Ciona intestinalis has been shown to be functional when expressed in cultured human cells (Hakkaart et al. 2006). Therefore, the quinol terminal oxidase activity of AOX has been demonstrated in at least one member of the kingdoms Animalia, Plantae, Fungi, Protista and Eubacteria.
Cellular localisation and structural modelling: AOX is a membrane-bound di-iron carboxylate protein
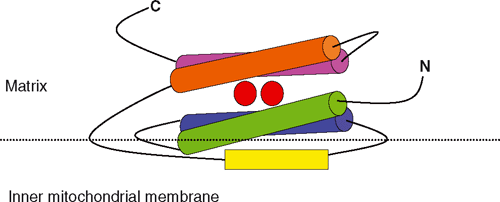
In order for AOX to be active as a quinol oxidase it must have access to its substrate and this requires interaction between AOX and a membrane where quinols are present. All AOX proteins that have been investigated to date are associated with membranes as demonstrated by heterologous expression studies in E. coli in which both prokaryotic and eukaryotic AOXs are recovered in the membrane fractions (Nihei et al. 2003; Stenmark and Nordlund 2003; Suzuki et al. 2004). Prior work has shown that there is a length difference between prokaryotic and eukaryotic AOX sequences; specifically that the N-termini of eukaryotic AOX proteins are much longer than their prokaryotic counterparts (Finnegan et al. 2003; Stenmark and Nordlund 2003; McDonald and Vanlerberghe 2006; Fig. 3). In eukaryotes AOX associates with the inner mitochondrial membrane (Day and Wiskich 1995; Vanlerberghe et al. 1998; Kirimura et al. 2006; Fig. 1). This subcellular localisation in soybean (and likely other eukaryotes) requires the presence of a presequence for mitochondrial targeting (Tanudji et al. 1999; Fig. 3), and provides an explanation as to why the N-terminal regions of eukaryotic AOXs are longer than those of prokaryotes (Fig. 3). Several eukaryotic AOX sequences that have been examined using the MITOPROT software (Claros and Vincens 1996) to predict the mitochondrial targeting sequence cleavage site contain this site 43 to 65 amino acids from the beginning of the protein, whereas no cleavage sites are predicted for prokaryotic AOXs (Fig. 3).
|
Bacterial AOXs might approximate the minimal unit or sequence length required for AOX activity based on the observation that bacterial AOXs lack most of the N-termini that are seen in the AOX sequences of eukaryotes, yet the AOX from the bacterium N. aromaticivorans is still active (Finnegan et al. 2003; McDonald et al. 2003; Stenmark and Nordlund 2003; Atteia et al. 2004; Fig. 3). The information responsible for the cell-membrane targeting of prokaryotic AOXs is either unnecessary as suggested by Finnegan et al. (2003) or must be contained within the minimal AOX unit because a lengthy N-terminal region is not necessary for the membrane targeting of N. aromaticivorans AOX.
AOX catalyses the four-electron reduction of oxygen to water and the identification of conserved amino acid motifs in the protein suggests the presence of a binuclear iron centre in contrast to COX, which contains heme (Siedow et al. 1995). AOX is proposed to consist of four helices that are associated with the inner mitochondrial membrane and this structure is likely to serve as an effective scaffold for the binding and activation of oxygen (Affourtit et al. 2002; Sazinsky and Lippard 2006; Fig. 4). The possibility that AOX might be a member of the di-iron carboxylate protein family has been suggested based on the presence of two putative iron-binding motifs containing conserved glutamate (E) and histidine (H) residues (E-X-X-H) and a model was put forward to identify the key residues important for the co-ordination of the di-iron centre (Siedow et al. 1995). Other members of this protein family include ribonucleotide reductase subunit R2, ferritins, bacterioferritins, the stearoyl-ACP Δ9-desaturase, rubrerythrins and methane monooxygenase (Berthold et al. 2000).
|
A structural model for AOX has been proposed using the known 3-D structures of other binuclear iron proteins as a guide (Moore et al. 1995; Siedow et al. 1995). As a result of hydrophobicity mapping, two transmembrane domains were thought to anchor each subunit into the membrane (Siedow et al. 1995). The model was later modified after taking into account expanded sequence and structural information for di-iron carboxylate proteins. Although the di-iron centre was retained, a reassignment was proposed for the E and H ligands (Andersson and Nordlund 1999). This second model was also supported by the newly available sequence data of several fungal AOXs (Joseph-Horne et al. 2000). In the new model, the transmembrane domains are no longer structurally tenable, and the hydrophobic part of the protein is believed to associate with the inner membrane, thereby making AOX an interfacial membrane protein (Andersson and Nordlund 1999; Affourtit et al. 2002; Fig. 4). This inner membrane associated region is thought to contain the ubiquinol binding site (Andersson and Nordlund 1999; Fig. 4). The above models have been refined as more sequence data have become available for AOX proteins from plants and other organisms (Berthold et al. 2000). Evidence from electron paramagnetic resonance spectroscopy was finally able to demonstrate that AOX did indeed possess a di-iron carboxylate centre and AOX is now classified as a member of the membrane-bound di-iron carboxylate proteins (Berthold et al. 2002; Berthold and Stenmark 2003; Moore et al. 2008).
Catalysis: residues important for AOX quinol oxidase activity
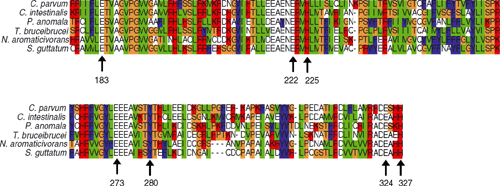
The advantage of the above models is that they make predictions about what residues in the AOX protein are involved in catalysis. These hypotheses are based on the structural model and the presence of highly conserved residues identified using multiple sequence alignments of AOX proteins from diverse organisms. The proposed iron ligands (using the numbering of the amino acid sequence of the Arabidopsis thaliana L. Heynh. AOX1a) are: E-183, E-222, H-225, E-273, E-324 and H-327 (Berthold et al. 2000; Fig. 5). The role of some of these residues in AOX activity has been tested using site-directed mutagenesis in plants and other organisms and studies have shown that several residues are required for activity (Table 1).
|
An attempt to identify the AOX residues involved in quinol binding was conducted based on the belief that SHAM competitively inhibits AOX by binding to the quinol binding site (Berthold 1998). Randomly mutated A. thaliana AOX proteins were expressed in a heme-deficient E. coli strain and four amino acid mutations located between the second and third helix were recovered that were resistant to SHAM inhibition, implying that they might play a role in quinol interactions (Berthold 1998). Other enzymes that react with quinols (e.g. bacterial photosynthetic reaction centres) contain a somewhat conserved motif: aliphatic residue-(X)3-H-(X)2/3-(L/T/S) that comprises residues 135 and 150 in some AOXs (Fisher and Rich 2000). Tyrosine (Y) radical formation might take place during oxidation reactions, electron transfers or quinol substrate interactions (Berthold et al. 2000). Y-280 has been hypothesised to play a role in such a reaction based on the fact that site-directed mutagenesis of this residue to phenylalanine (F) yielded an inactive AOX, while the mutation of other conserved Y residues did not impact AOX activity (Fig. 5; Table 1). It is proposed that the presence of a hydroxyl moiety in this location is important as Y radicals can be involved in reduction and oxidation mechanisms involving oxygen (Berthold et al. 2000; Affourtit et al. 2002).
Other conserved residues, such as L-182, A-186, N-221, A-276 and D-323, are near the proposed iron-binding residues (Fig. 5). The conservation of these residues might simply result from their proximity to iron-binding residues, or they might play a more direct role in AOX catalysis, regulation or structure. N221 and D323, for example, are proposed to hydrogen bond to H327 and H225, respectively, and might function to stabilise the helical structure of AOX (Berthold et al. 2000). Surprisingly, E-183, which is proposed to be an iron-binding ligand, has not been investigated using site-directed mutagenesis to confirm whether it is necessary for AOX activity.
How are AOX gene expression and AOX protein activity regulated?
How many AOX genes are present in a genome?
AOX is a nuclear-encoded gene in eukaryotes and occurs as a multigene family in many plants, including soybean, A. thaliana, tobacco, corn, mango, sugarcane, wheat, cotton and rice (Whelan et al. 1995, 1996; Saisho et al. 1997; Considine et al. 2001; Karpova et al. 2002; Saika et al. 2002; Takumi et al. 2002; Borecky et al. 2006; Li et al. 2008). In angiosperms, two types of AOX genes exist: AOX1 genes have been found in all angiosperms examined to date, while AOX2 genes appear to be limited to dicot speces (Considine et al. 2002). The physiological significance of this fact remains to be determined, but it has been proposed that some AOX genes may be expressed in order to carry out ‘housekeeping’ functions, whereas others might play a role in a response to specific stresses (Considine et al. 2002).
AOX is also present as a multigene family in the fungus Candida albicans (Huh and Kang 2001) and homology searches have identified more than one AOX sequence in Botrytis cinerea, Candida maltosa, Candida tropicalis, Coccidioides posadasii and Podospora anserina (McDonald and Vanlerberghe 2006), which might result from several AOX genes being present in these genomes. Several protists contain more than one AOX gene, such as C. reinhardtii (Dinant et al. 2001), Perkinsus marinus, Tetrahymena thermophila, Thalassiosira pseudonana and Euglena gracilis (McDonald and Vanlerberghe 2006). To date, no examples of a eubacterium or an animal with more than one AOX gene have been identified (McDonald and Vanlerberghe 2006). Thus, in some organisms, experiments are complicated by the presence of an AOX multigene family and care must be taken to develop gene-specific probes, which are usually designed based on the 5′ or 3′ untranslated regions (Ito et al. 1997).
Transcriptional control of AOX genes
Because of the position of the AOX protein in the respiratory electron transport chain (ETC) and its influence on energy generation it is clear that AOX gene expression must be under tight control. In prokaryotes, AOX expression has only been examined in the eubacterium Novosphingobium aromaticivorans and is affected by oxygen level and the carbon source in the growth media (Stenmark and Nordlund 2003). In eukaryotes, AOX is encoded by nuclear genes, but the protein is localised to the inner mitochondrial membrane. Therefore, AOX gene expression must be influenced to some degree by the metabolic state of the mitochondrion. In the case of AOX, this occurs through the process of mitochondrial retrograde regulation (MRR), whereby changes in nuclear gene expression are directed by the mitochondrion (Rhoads and Subbaiah 2007). The pathways involved in transferring these signals from the mitochondrion to the nucleus are a rapidly expanding area of research and AOX has played a pivotal role in these studies (Rhoads and Subbaiah 2007).
AOX expression is affected by a variety of biotic and abiotic stresses. This has been demonstrated by many studies using northern blots, real-time PCR or microarray technologies to examine patterns of gene expression under a wide variety of experimental and environmental conditions. AOX1 gene expression in tobacco and soybean can be influenced by intermediates of the tricarboxylate acid cycle; in particular, citrate, malate and 2-oxoglutarate (Vanlerberghe and McIntosh 1996; Djajanegara et al. 2002; Gray et al. 2004). This is not particularly surprising given that the reducing equivalents generated at several steps of the cycle are used by dehydrogenases to input energy into the mitochondrial ETC. The metabolites acetate and cysteine can also increase AOX1 expression in tobacco (Vanlerberghe and McIntosh 1996). Inhibitors of respiratory complexes also influence AOX gene transcription. The complex III (cytochrome c oxidoreductase) inhibitor antimycin A increases the expression of AOX1 in tobacco (Vanlerberghe and McIntosh 1996), soybean AOX1 (Djajanegara et al. 2002) and AOX1a in A. thaliana, but not AOX1b, AOX1c or AOX2 expression (Saisho et al. 1997). The complex IV (COX) inhibitor potassium cyanide (KCN) increases the expression of corn AOX1a (Polidoros et al. 2005), whereas the cytochrome bc1 complex inhibitor SSF-126 increases AOX expression in the fungus Magnaporthe grisea (Yukioka et al. 1998). In the fungus Neurospora crassa aod-1 (AOX) gene expression increased dramatically in strains that were deficient in cytochromes (Li et al. 1996).
Compounds associated with oxidative stress also influence AOX transcript levels. Reactive oxygen species (ROS) generating treatments increased AOX1a expression, but repressed AOX2 expression in Arabidopsis cell cultures (Clifton et al. 2005). The application of H2O2 increases AOX1 expression in tobacco (Vanlerberghe and McIntosh 1996), AOX1a expression in corn (Polidoros et al. 2005) and AOX expression in the fungus Magnaporthe grisea (Yukioka et al. 1998). The ROS generator paraquat is capable of increasing AOX expression in the fungus Aspergillus fumigatus (Magnani et al. 2007).
In addition, several systemic signalling molecules influence AOX expression. Salicylic acid increases AOX1 expression in soybean (Djajanegara et al. 2002), methyl salicylate increases AOX expression in tomato (Fung et al. 2006), and ethylene and jasmonic acid increase AOX1a expression in tobacco (Ederli et al. 2006).
Several environmental factors influence AOX expression. Low temperature (4–10°C) leads to an increase in the expression of AOX1a and AOX1b in rice and soybean AOX1 (Ito et al. 1997; Djajanegara et al. 2002). Osmotic stress in cowpea and ozone treatment of tobacco have also been shown to influence AOX expression (Ederli et al. 2006; Costa et al. 2007). Expression of AOX1a in A. thaliana is increased by treatment of plants with ozone, salt, ultraviolet light, cold and osmotic stress (Elhafez et al. 2006). Work has been conducted in C. reinhardtii to examine the influence of nitrogen sources (ammonium and nitrate) on AOX transcripts (Baurain et al. 2003).
Different AOX genes show different expression patterns both spatially and temporally. In A. thaliana, AOX1a and AOX1c are detected in the flowers, buds, stems, rosettes and roots, AOX1b is seen in flowers and buds, and AOX2 is detected in stems, rosettes and roots (Saisho et al. 1997). This work was further supported in A. thaliana by an examination of all AOX transcripts by conducting a meta-analysis of microarray gene chip data that examined the expression of the genes in many tissues and developmental stages (Clifton et al. 2006). In Trypanosoma brucei the life-cycle stage also affects AOX expression (Chaudhuri et al. 2002).
Although a great deal is known about the influence of various stresses on AOX gene expression, less is known about how these signals are relayed to the transcription machinery and the influence of cis and trans regulatory elements on AOX gene transcription. Analysis of the promoter regions of AOX genes in fungi and plants is progressing and several positive and negative regulatory elements have now been identified.
The first plant AOX promoter region to be analysed was from the AOX1 gene of S. guttatum (Rhoads and McIntosh 1993). The promoter contains a putative TATA box and several cis-acting transcriptional elements, among them putative recognition sequences for zinc finger and basic leucine zipper proteins (Rhoads and McIntosh 1993). Regions of the AOX1 promoter also displayed sequence similarities with promoter regions from PR1a (Pathogenesis-Related) and GRP8 (Glycine Rich Protein) genes, which are involved in responses to pathogen attacks (Rhoads and McIntosh 1993).
Examination of the AOX1 promoter in C. reinhardtii demonstrated that nucleotides −253 to +59 (relative to the transcription start site) are sufficient for gene expression and regulation, but that other elements were required for full gene expression (Baurain et al. 2003). Although AOX1 expression is stimulated by nitrate, a nitrogen-related element was not identified in the promoter (Baurain et al. 2003).
It has been known for many years that members of multigene families of AOX in plants are differentially regulated (Ito et al. 1997). An analysis of the promoter regions of the AOX1 and AOX2b promoters from soybean indicated that both positive and negative regulatory elements exist within 2 kB upstream of the translational start; this analysis also demonstrated that additional elements must be present further upstream of this region, or that some elements might be located in the 3′ end of these genes (Thirkettle-Watts et al. 2003). A comparison of the promoters of AOX genes from A. thaliana and soybean revealed the presence of several common motifs, including elements or binding sites believed to play a role in tissue or development specificity and responses to environmental cues (Thirkettle-Watts et al. 2003). An analysis of the promoter region of the AOX1c gene of A. thaliana using various deletion constructs revealed that positive and negative response regions and elements are present (Ho et al. 2007). Seven sequence elements of interest were also identified through a comparison of the AtAOX1c promoter with the promoter of the soybean GmAOX2b gene (Ho et al. 2007). Several of these elements were found to be functional in the AtAOX1c promoter and were also recognisable in other plant promoter regions from Oryza sativa L., Zea mays L., S. guttatum, Lotus corniculatus L. and Catharanthus roseus L. G. Don, indicating that there might be some conservation of regulation in some plant AOX gene promoters (Ho et al. 2007).
In terms of identifying transcription factors that interact with an AOX gene promoter, the most progress in this area has occurred in the fungus Neurospora crassa, where five genes (aod-2, aod-4, aod-5, aod-6 and aod-7) required for aod-1 (AOX) gene expression have been isolated (Descheneau et al. 2005). Recently, by generating mutations in the upstream region of the aod-1 gene and by growing the cells on antimycin A (an inhibitor of complex III) an AOX induction motif was identified (Chae et al. 2007a). It was hypothesised that the motif, which consists of a pair of CGG repeats separated by 7 bp, might be recognised by a member of the zinc-cluster family of transcription factors that might activate aod-1 expression by binding to the sequence (Chae et al. 2007a). Indeed, the aod-2 and aod-5 gene products have recently been identified as transcription factors of this family (Chae et al. 2007b). It is worth noting that the induction motif present in the aod-1 gene of N. crassa was only conserved in other members of the fungal order Sordariales. This indicates that the transcriptional regulation pathways of individual AOX genes may very well be gene specific and might differ not only between AOX genes in a single species (i.e. within multigene families), but also between AOX genes from different organisms representing diverse kingdoms.
Post-translational regulation of AOX enzyme activity
Subunit structure: monomeric versus dimeric
A major objective of current research is to understand the biochemical mechanisms by which the AOX enzyme is regulated via fine metabolic control because this is likely to have a major impact on the in vivo partitioning of electrons to AOX. It is important to distinguish between AOX capacity and AOX engagement. AOX capacity is the maximum AOX activity of a cell or tissue, or an estimate of the maximum possible flux of electrons to AOX. It is measured by the addition of a cyt pathway inhibitor (such as CN) followed by the addition of an AOX inhibitor (such as nPG). Capacity is measured under these conditions as the O2 uptake resistant to the cyt pathway inhibitor and sensitive to the AOX inhibitor (Møller et al. 1988). AOX capacity is not indicative of the actual flux of electrons to AOX in the cell before the introduction of the inhibitors.
AOX engagement involves the estimation of the actual flux of electrons to the AOX pathway under physiological conditions and is, therefore, much more difficult to measure than AOX capacity. The best way to measure engagement is by using a non-invasive oxygen isotope discrimination technique (Guy et al. 1989). This method is based on the fact that AOX and COX discriminate against the isotope 18O to different extents. Both gas-phase (for use with whole tissue) and aqueous-phase (for use with mitochondria or cell culture) systems connected online to a gas chromatography-mass spectrometry unit have been used for this purpose (Robinson et al. 1995). The sample is placed in a leak-tight reaction vessel from which gas samples can be withdrawn at regular intervals as oxygen is consumed by respiration. To determine a discrimination value, both the oxygen concentration and its isotopic ratio are then determined for each sample. These values are then compared with the end-point discrimination values for AOX and COX, which are determined separately for each pathway after chemical inhibition of the other pathway.
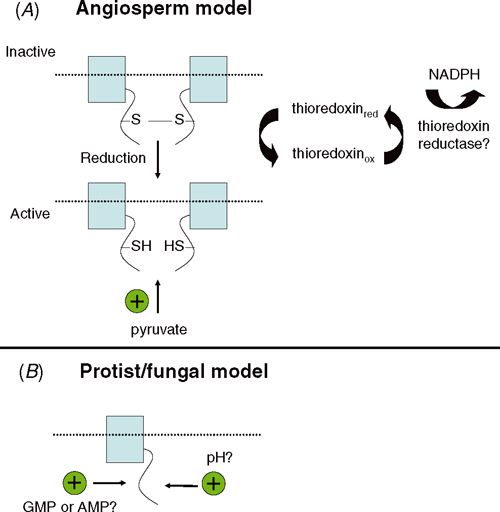
AOXs from several protists and fungi appear to be monomeric (Table 2). In contrast, angiosperm AOX is a covalently linked dimer that consists of identical or similar subunits (possibly encoded by different AOX genes) that are linked in a reversible manner by a disulfide bond (Umbach and Siedow 1993; Table 2). Isolation of mitochondria from the leaves of AOX overexpressor lines demonstrated that AOX protein can be present as an oxidised or reduced dimer (Vanlerberghe et al. 1995).The relative abundance of the oxidised and reduced forms can be evaluated by a combination of reducing and non-reducing SDS–PAGE and immunoblot analysis (Umbach and Siedow 1993). The oxidised tobacco AOX is ~70 kDa in size and the reduced AOX is ~35 kDa in size using non-reducing SDS–PAGE (Vanlerberghe et al. 1998). The dimer, when covalently linked by a disulfide bond between the two subunits, is a less active form of AOX (as determined by in organello assays), while reduction of the disulfide bond to its component sulfhydryls produces a more active form (Umbach and Siedow 1993; Fig. 6A).

|
|
Alternative pathway capacity is higher in soybean mitochondria when AOX is present in a reduced form compared with an oxidised form, indicating that the sulfhydryl/disulfide interconversion can affect electron flow through the pathway (Umbach and Siedow 1993). AOX can be interconverted from the oxidised to the reduced form by the sulfhydryl reductant dithiothreitol (DTT), and from the reduced to oxidised form by the sulfydryl reagent azodicarboxylic acid bis (dimethyl-amide) (diamide) (Umbach and Siedow 1993). In other words, there is a redox modulation of AOX activity by sulfhydryl/disulfide interconversion (Fig. 6A).
The amino acid cysteine contains a sulfhydryl group that can participate in this type of reaction and considereable focus has been directed to two highly conserved cysteine (Cys) residues in the N-terminal hydrophilic domain of most angiosperm AOXs (Fig. 3). The tobacco AOX1 cDNA has two cysteines (Cys) at positions 126 (CysI) and 176 (CysII), which are predicted to reside in the mitochondrial matrix and might be responsible for the redox regulation and/or activation of AOX. The Cys residue responsible for this regulation has been determined using site-directed mutagenesis (Rhoads et al. 1998; Vanlerberghe et al. 1998). Mutation of Cys-126 to an alanine resulted in an AOX that could not form a disulfide bond, indicating that CysI is responsible for the intersubunit disulfide bond formation (Vanlerberghe et al. 1998).
Reduction of AOX can also occur quickly in response to mitochondrial metabolism (Vanlerberghe et al. 1995). This reduction event can also be mediated by the in vivo oxidation of citrate, isocitrate and malate (Vanlerberghe et al. 1995). Work with transgenic tobacco plants that overexpress the mitochondrial isoform of NADP+-dependent isocitrate dehydrogenase (mtICDH) sevenfold has shown an increase in the reductive activation of AOX compared with controls (Gray et al. 2004). Citrate feeding of the transgenic plants led to a high conversion of AOX from the oxidised to the reduced form (Gray et al. 2004). These authors hypothesised that mtICDH could regulate flux through the tricarboxylic acid (TCA) cycle and could also influence AOX activity through modulation of its reduction status (Gray et al. 2004). This NADP+ specificity suggests that NADPH might be required for AOX reduction and that the reduction might be mediated by a mitochondrial thioredoxin or glutathione system (both of which specifically require NADPH) (Vanlerberghe et al. 1999; Fig. 6A).
A mitochondrial-specific thioredoxin isoform PtTRXh2 exists in plants and gives credence to this possibility (Laloi et al. 2001; Gelhaye et al. 2004). This thioredoxin could be effectively reduced by a mitochondrial thioredoxin reductase and is capable of reducing AOX in organello, indicating that the conversion of AOX from the oxidised to the reduced form might be mediated by a thioredoxin system (Gelhaye et al. 2004; Fig. 6A). This reduction occurs quite rapidly, which implies that the sulfhydryl/disulfide system can provide short-term fine control for the regulation of AOX activity.
In addition to the sulfhydryl/disulfide regulatory system, angiosperm AOX activity is strongly dependent on the presence of particular α-keto acids, most notably pyruvate, but also including glyoxylate, hydroxypyruvate and 2-oxoglutarate (Millar et al. 1993; Fig. 6A). Pyruvate activation takes place from within the mitochondrial matrix, is fully reversible, and is not dependent on pyruvate metabolism (Millar et al. 1993). Furthermore, only the more active, reduced form of AOX is subject to pyruvate activation (Umbach et al. 1994; Vanlerberghe et al. 1995; Fig. 6A). Pyruvate acts to increase the maximum rate (Vmax) of the AOX reaction, possibly by preventing inhibition of the enzyme by oxidised ubiquinone (Hoefnagel and Wiskich 1998). Early studies also suggested that pyruvate action resulted from its interaction with a Cys sulfhydryl to form a thiohemiacetal because activation was mimicked by iodoacetate (Umbach and Siedow 1996; Fig. 6A). Mutation of CysI results in a large drop in AOX activity and in a loss of activation by pyruvate; however, the CysII mutant is not affected (Vanlerberghe et al. 1998). This indicates that CysI is required for AOX activation by pyruvate. It remains to be determined whether the mitochondrial pyruvate concentration is sufficient in vivo to regulate AOX in this fashion.
Therefore, significant AOX activity in tobacco mitochondria is dependent on both the reduction of the regulatory disulfide bond and on the presence of pyruvate (Vanlerberghe et al. 1995; Fig. 6A). However, experimental work in durum wheat mitochondria has demonstrated that AOX in this species is powerfully activated by the photorespiratory cycle intermediates hydroxypyruvate and glyoxylate, but unlike other plant AOXs durum wheat AOX was not activated by 2-oxoglutarate (Pastore et al. 2001). In addition, when CysI is mutated to serine the enzyme cannot be oxidised and is specifically activated by succinate (Djajanegara et al. 1999). These results indicate that there might be some species-specific variation in plant AOX post-translational regulation.
In contrast to the situation in angiosperms, the monomeric AOXs of other eukaryotes are post-translationally regulated in a different manner. Many of these AOXs are strongly stimulated by purine nucleoside 5′ monophosphates, especially GMP and/or AMP, as is the case for the fungi Moniliella tomentosa, N. crassa and Yarrowia lipolytica (Hanssens and Verachtert 1976; Vanderleyden et al. 1980; Medentsev et al. 2004; Fig. 6B). This effect is also seen in the protists Acanthamoeba castellani and D. discoideum (Jarmuszkiewicz et al. 2002, 2005). Several of these AOXs appear to be insensitive to stimulation by organic acids, in direct contrast to the angiosperm enzymes (Jarmuszkiewicz et al. 2002).
The optimum pH for AOX pathway activity in A. castellani is 6.8, which is lower than the pH optimum of 7.4 observed for the cyt pathway (Jarmuszkiewicz et al. 2002; Castro-Guerrero et al. 2004). This difference in the pH optima of the two pathways has also been seen in E. gracilis (Castro-Guerrero et al. 2004). It has been hypothesised that pH might represent an effective way to control AOX activity when the cyt pathway is impaired because the matrix pH is lower when the proton-pumping complexes of the cyt pathway are inactive and would, therefore, favour the AOX pathway (Jarmuszkiewicz et al. 2002; Fig. 6B).
What is the physiological function(s) of AOX?
As our understanding of AOX has developed at the biochemical and molecular levels, so too have hypotheses as to why organisms possess this respiratory pathway given the wasteful nature of AOX in terms of energy conservation. A quick survey of the primary literature reveals that AOX expression and AOX pathway capacity often increase in plants and other organisms during periods of biotic and abiotic stress (Juszczuk and Rychter 2003; Borecky et al. 2006; Castro-Guerrero et al. 2008). It is likely that AOX conveys metabolic flexibility to the respiratory ETC and that this provides an advantage to organisms that are subject to frequent and various types of environmental stress, particularly under conditions that limit the effectiveness of the cyt pathway. The ultimate role of the AOX pathway remains to be confirmed, but several hypotheses abound as to why it exists and its physiological function.
Thermogenesis
One demonstrated physiological function of AOX occurs during the process of thermogenesis in some plants. Recent work in the sacred lotus (Nelumbo nucifera Gaertn.) has shown that increased flux through the AOX pathway is responsible for heating during thermogenesis (Watling et al. 2006). Work in the dead horse arum (Helicodiceros muscivorus Engl.) demonstrated that thermogenesis results in heat and odour release from the plant and lures the blowfly pollinator to the plant to facilitate pollination (Angioy et al. 2004). Although this function explains the presence of AOX in thermogenic organs, it does not provide a rationale for why AOX is so widely expressed in non-thermogenic plant tissues and non-thermogenic organisms.
Balancing carbon metabolism and electron transport
Over-reduction of an ETC could conceivably occur when the electrons entering the ETC and the electrons leaving the ETC are not balanced. This might involve an increase in the activity of the enzymes capable of putting electrons into the ETC via the quinol pool (e.g. complex I, the alternative internal and/or external NAD(P)H dehydrogenases, succinate dehydrogenase (SDH) or other enzymes such as sulfide : quinone oxidoreductase). Alternatively, over-reduction of respiratory components could occur because of a decrease in the activities of the AOX and/or cyt pathways and a problem with the exit of electrons from the ETC. Therefore, over-reduction of the respiratory ETC could occur when large amounts of carbon are moving through glycolysis and the TCA cycle, during conditions where there is an increase in the amount of NAD(P)H being produced via various cellular processes, or under conditions where the AOX and/or cyt pathways are impaired.
In fact, one of the initial hypotheses about AOX function was that it acted as an overflow for the cytochrome pathway if too many electrons were entering the ETC (Lambers 1982). Subsequent work has illustrated that this is not the case and that the AOX pathway can in fact compete with the cyt pathway for respiratory electrons (Day et al. 1996). AOX antisense and overexpressor lines were also used to demonstrate that the AOX pathway is sufficient to support respiratory carbon metabolism under conditions that inhibit the cyt pathway (Vanlerberghe et al. 1997). AOX allows for the continuation of electron transport if the cyt pathway is limited by the supply of inorganic phosphate and ADP and allows TCA cycle turnover when the energy charge of the cell is high (Affourtit et al. 2002; Millenaar and Lambers 2003). This allows for the continued provision of carbon skeletons for growth and repair. Therefore, AOX might play a role in balancing carbon metabolism and electron transport, especially in organisms such as Novosphingobium aromaticivorans that can use a variety of compounds as energy and carbon sources (Stenmark and Nordlund 2003). AOX might be important in proteobacteria that are subjected to constantly changing environmental conditions and must therefore adapt their energy metabolism depending on the circumstances in which they find themselves. AOX might represent a means of allowing metabolic flexibility that is very important during times of stress and might act as a rheostat for the cell that monitors carbon flow and energy production based on the inputs and outputs of the system, perhaps via mitochondrial retrograde regulation (Rhoads and Subbaiah 2007).
AOX might serve to regenerate oxidised reductants (NAD+ and NADP+) and thereby prevent the inhibition of carbon metabolism under conditions of cyt impairment (Millenaar and Lambers 2003; Stenmark and Nordlund 2003). The ability to reoxidise NADH has been put forward as a possible function in C. parvum because this organism appears to lack the enzymes necessary for the TCA cycle and lacks a cyt pathway (Roberts et al. 2004).
Recent work has demonstrated that in plants AOX can be used to remove excess reducing power from the chloroplast. In an A. thaliana mutant lacking cyclic electron flow around photosystem I reducing equivalents will accumulate in the chloroplast stroma, but must be removed for efficient photosynthesis to occur (Yoshida et al. 2007). Results obtained with this mutant indicate that these reducing equivalents are transported from the chloroplast to the mitochondria where AOX serves to dissipate the excess reducing equivalents (Yoshida et al. 2007). This is proposed to occur by shunting the reducing equivalents via the oxaloacetate/malate shuttle to the mitochondrion where they are taken up by the ETC and dissipated via mitochondrial non-phosphorylating pathways (including AOX) (Noguchi and Yoshida 2008). Durum wheat mitochondria might also prevent chloroplast or cytosol over-reduction by using the alternative rotenone-insensitive external NAD(P)H and the malate/oxaloacetate transporter to move reducing equivalents into the mitochondria during times of stress where AOX is active because of its activation by intermediates of the photorespiratory cycle (Pastore et al. 2007).
Control of reactive oxygen species generation
AOX might influence the generation of ROS by the respiratory chain (Maxwell et al. 1999; Møller 2001). Mitochondrial ETCs are a significant source of ROS and the rate of ROS generation is dependent on the reduction state of key respiratory components. These mitochondrial ROS might have important signalling functions within cells, but their excessive generation may cause oxidative damage to the mitochondrion. By preventing the over-reduction of the respiratory chain, AOX could act to reduce ROS generation. As AOX is catalytically active as a monomer or dimer, and can likely be produced faster than multi-subunit complexes, it has been proposed as a means of using any excess electrons in the ETC, thus preventing a great deal of ROS production. In support of this hypothesis, transgenic plant cells lacking AOX display higher rates of mitochondrial ROS generation, particularly under growth conditions that might promote a more reduced chain (Maxwell et al. 1999). Activation of the AOX of A. castellani lowered H2O2 production, but this effect was cancelled by benzohydroxamate, leading the authors to propose that AOX was protecting the organism against mitochondrial oxidative stress (Czarna and Jarmuszkiewicz 2005). In Ustilago maydis, ROS production is increased in the presence of the cyt inhibitor CN, but this effect is lower in cells showing high AOX pathway capacity (Juárez et al. 2006). An increase in AOX capacity in response to H2O2 has been observed in E. gracilis, suggesting that it is part of a cellular response to oxidative stress (Castro-Guerrero et al. 2004). Fasting in the amoeba Chaos carolinensis also increases oxidative stress and the activation of AOX is seen as a protective mechanism (Deng et al. 2002). In A. fumigatus AOX mRNA expression is induced by paraquat and menadione, two compounds that lead to cellular oxidative stress (Magnani et al. 2007). AOX activation in durum wheat mitochondria also led to a decrease in the rate of superoxide anion generation (Pastore et al. 2001). In cold and high-oxygen-treated bell pepper and cauliflower AOX is induced and superoxide production is lower than in the control (Popov et al. 2001). The above examples argue for a role of AOX in the prevention of oxidative stress and/or a response to oxidative stress.
Much of the recent work in fungi has centred on the role of AOX and how reducing ROS generation may lead to an extended lifespan. Research in Podospora anserina has demonstrated that an enhancement in the expression of AOX leads to an increase in lifespan, most likely because of a decrease in the generation of ROS (Gredilla et al. 2006). AOX levels have also been seen to change depending on the life-cycle stage of an organism. The activity and protein levels of AOX decrease when cells of A. castellani shift from the exponential growth phase to the stationary phase (Czarna et al. 2007). AOX levels in D. discoideum were high during exponential growth and decreased after entry into the stationary phase of growth (Jarmuszkiewicz et al. 2002).
O2 scavenging
Excess O2 can be dangerous because it can lead to the generation of ROS. Extant di-iron carboxylate proteins catalyse a variety of reactions, but studies have demonstrated that many of these enzymes are also capable of using O2 as a substrate. Diverse members of this protein family retain a capacity to reduce O2 to water, even though these same proteins are thought to primarily have other biochemical functions (Broadwater et al. 1998; Gassner and Lippard 1999; Gomes et al. 2001). Based on this evidence it has been suggested that the original function of di-iron carboxylate proteins was to act as oxidases (Gomes et al. 2001). Di-iron carboxylate proteins are found in all kingdoms of life and are, therefore, likely to be of ancient origin. It has been proposed that the evolutionary driving force for the improved oxidase function of AOX might have been the transition from an anoxic world to one in which toxic O2 levels were starting to rise (McDonald and Vanlerberghe 2006). AOX is sulfide resistant (Azcón-Bieto et al. 1989) and, therefore, would have been able to operate in the ancient sulfide-rich world when perhaps cyt could not (McDonald and Vanlerberghe 2006). The original role of AOX might have been that of an O2-scavenging enzyme and this function might have been retained in some lineages in which it is present today.
AOX might be an evolutionary adaptation to O2 stress conditions in the protistan parasite C. parvum (Suzuki et al. 2004). This organism lives best in a low O2 intestinal environment and AOX might represent a means of removing localised excess O2 (Putignani et al. 2004). A similar role in marine molluscs has been postulated during metabolic rate reduction, ADP limiting conditions or while the cyt pathway is O2 saturated as a means of maintaining the intracellular partial pressure of O2 (PO2) (Abele et al. 2007). In contrast, the AOX of N. aromaticivorans is unlikely to play a role in O2 scavenging because it is expressed at higher levels at lower O2 concentrations (Stenmark and Nordlund 2003).
Resistance to metals, toxins and poisons: role in pathogenicity
One of the original hallmarks of AOX respiration is its ability to continue in the presence of the respiratory poison CN in contrast to the cyt pathway. The AOX pathway might serve as an alternative system that uses electrons in place of the incapacitated cyt pathway, thereby allowing glycolysis and respiration to continue to meet the needs of the organism. In support of this hypothesis, loss of the cyt pathway (by chemical or genetic means) results in an induction of AOX expression in plants, fungi and protists (Djajanegara et al. 2002; Castro-Guerrero et al. 2004; Juárez et al. 2004).
Research over the years has demonstrated that AOX proteins are insensitive to many compounds and molecules that effectively inhibit COX (complex IV) or complex III. These inhibitors include sulfide (Azcón-Bieto et al. 1989), cadmium (Castro-Guerrero et al. 2008), nitric oxide (NO) (Huang et al. 2002), azide (Baurain et al. 2003), antimycin A (Chae et al. 2007a) and myxothiazol (Cournac et al. 2002). These inhibitors might accumulate in tissues as a result of their abundance in particular environments or simply as products of normal physiological and biochemical processes. This resistance of AOX to a wide spectrum of hazardous compounds might be especially advantageous for microorganisms such as bacteria, fungi and protists that are often involved in competition with other organisms for limited resources in their respective biological niches.
Results indicate that the presence of AOX is likely to be widespread in the animal kingdom (McDonald and Vanlerberghe 2006). Many of the animals that possess AOX live in marine sediments where high concentrations of sulfide might inhibit COX (Nicholls 1975). Invertebrates in these habitats display elevated sulfide resistance as a result of several physiological and metabolic adaptations (Grieshaber and Völkel 1998). Mitochondria from marine invertebrates can oxidise sulfide to thiosulfate and can pass the electrons to oxygen via the respiratory chain, perhaps via a sulfide : quinone oxidoreductase (SQR) that is present in many animals (Grieshaber and Völkel 1998; Theissen et al. 2003). Under low-sulfide conditions, electrons from sulfide oxidation and/or TCA cycle reactions could be passed to oxygen via the usual cyt pathway and coupled to ATP production. However, at higher sulfide concentrations (when cyt is inhibited by sulfide), it has been hypothesised that electrons are passed to oxygen by another terminal oxidase, allowing aerobic metabolism and the continued detoxification of sulfide, thus promoting survival of the animal (Grieshaber and Völkel 1998). The characteristics of this oxidase are reminiscent of AOX, that is, CN-resistant, SHAM-sensitive and less tightly coupled to ATP production (Grieshaber and Völkel 1998). Therefore, AOX might play an important role in the metabolism of animals that live in or near hydrothermal vents where sulfide levels are extremely high (e.g. Riftia pachyptila).
AOX appears to play a role in resistance to metal toxicity in the protist E. gracilis (Castro-Guerrero et al. 2008). Although cadmium has been found to decrease the activities of complexes III and IV, AOX was relatively resistant to this inhibition (Castro-Guerrero et al. 2008). During exposure to cadmium, AOX activity made up 69–91% of the total respiration in E. gracilis (Castro-Guerrero et al. 2008). The authors hypothesise that AOX is a key component of a cadmium-resistance mechanism and that it allows for maintenance of the cell’s energy status during times of stress (Castro-Guerrero et al. 2008).
AOX might increase the pathogenicity of some organisms because of the fact that host defence responses are often characterised by the generation of reactive species (e.g. nitric oxide) or poisons (CN) in an attempt to fight off invaders. More than 40 pathogenic species from five kingdoms of life were found to contain AOX (McDonald and Vanlerberghe 2006); consistent with the hypothesis that AOX might play some role in pathogenesis, perhaps by rendering such organisms less sensitive to various cyt inhibitors.
AOX has been found to play a role in the pathogenesis of the fungus Cryptococcus neoformans, perhaps by improving its survival within phagocytic host cells (Akhter et al. 2003). Trypanosoma brucei brucei (the causative agent of sleeping sickness) uses AOX during its life-cycle stage in the blood stream of mammals and relies on AOX to reoxidise NADH and perhaps to prevent the generation of ROS (Fang and Beattie 2003).
Recently, promising results have been obtained using the AOX inhibitor SHAM to inhibit the growth of the apicomplexan parasites C. parvum and Toxoplasma gondii (Roberts et al. 2004). Work using novel AOX inhibitors is progressing well in T. brucei brucei (Ott et al. 2006). Therefore, basic research has led to the identification of AOX as a promising target for drug therapy. This work could also be extended to other pathogenic organisms that contain AOX, such as opportunistic fungal or protist pathogens (e.g. Candida albicans and Prototheca wickerhamii).
Role in cellular reprogramming
It is clear that AOX plays a significant role in an organism’s response to environmental change. An overarching role for AOX as a mediator of a coordinated response to changing conditions has recently been proposed (Clifton et al. 2006). In this case, not only would AOX act as a terminal oxidase, but it would also initiate cellular reprogramming appropriate to the environmental situation (Clifton et al. 2006). It is for this reason that AOX has been suggested as a possible genetic marker that could be used to recognise that efficient cellular reprogramming is taking place during exposure to a given stress (Arnholdt-Schmitt et al. 2006).
Future directions
Most progress to date in the field of AOX has come from studies conducted on relatively few organisms. Impressive work has been carried out in plants (e.g. Nicotiana tabacum L., Glycine max L. Merr., O. sativa), fungi (e.g. N. crassa, C. albicans) and protists (e.g. C. parvum, D. discoideum, T. brucei) (see Joseph-Horne et al. 2000; Jarmuszkiewicz et al. 2002; Finnegan et al. 2004; Chaudhuri et al. 2006 for an overview of some of these areas). The majority of research on AOX has been performed in the context of an assumption that AOX has a limited taxonomic distribution and that its presence in an ETC is the exception rather than the rule. This assumption might influence the questions that we think to ask about AOX; in effect introducing a bias. Public, on-line gene databases have led to the discovery that AOX is present in the genomes of organisms as varied as single-celled eubacteria to complex animals (McDonald and Vanlerberghe 2006). This represents a paradigm shift in the way that we think about AOX and the previous work conducted in plants, fungi and protists must now be placed into a much broader context. The question is no longer why only a few organisms contain AOX, but rather why do such a large number of biologically diverse organisms have AOX? Taxonomically speaking, AOX research to date has only examined the tip of the iceberg. Little is known about the AOXs of the majority of organisms in Fig. 2, except that the sequences exist, and there are obviously huge research opportunities available for exploration. Increased dialogue between researchers that are working on AOX in different organisms will provide the advantage of a comparative approach that will yield valuable insights into AOX function and regulation.
The genomic and post-genomic eras have provided scientists with new tools to explore questions about AOX. Exploration into the taxonomic distribution of AOX has led to the identification of novel AOX-containing organisms and to a huge increase in the amount of AOX sequence information available for analyses. One of these novel AOX-containing organisms might represent an ideal model system in which to ask particular questions about AOX function and regulation. The large amount of AOX sequence information can be used to identify conserved amino acids within the AOX protein that are logical candidates for site-directed mutagenesis work. Functional and regulatory hypotheses involving the conserved glutamate and histidine residues, putative quinol binding region, and other residues important for catalysis can be explored using the many AOX sequences now available from a range of diverse organisms.
The heterologous expression studies discussed in this review demonstrate that AOXs from a wide variety of kingdoms are functional quinol oxidases and that several of the AOX sequences recovered by database searches and PCR technologies have been shown to be true and active quinol oxidases. However, it is also necessary to determine whether AOX sequences recovered through bioinformatics approaches are biochemically active as quinol terminal oxidases in vivo. A related question is whether AOX can utilise only ubiquinol, or if other quinols can serve as substrates depending on the conditions and/or the species. AOX is found in a wide variety of organisms that are no doubt capable of making a large array of quinol compounds.
Improved technologies and isolation techniques allow for better mitochondrial and protein preparations that are being used to explore questions about AOX enzyme activity and post-translational modification of the AOX protein. To date, two modes of post-translational regulation have been identified. In general, the AOXs of fungi and protists are monomeric and subject to regulation by GMP and/or AMP, whereas those of angiosperms are dimeric and subject to redox regulation and pyruvate activation. These organisms exhibit divergent evolutionary histories. The fact that both D. discoideum and fungal AOXs are monomeric might indicate that it is likely that the AOXs of icthyosporeans, choanoflagellates and animals will also be monomeric (Fig. 2). It also seems likely that bacterial AOX will be monomeric and this might represent the ancestral state of the enzyme. It is quite possible that a dimeric AOX structure is a derived state found only in some plants. These hypotheses can be tested by examining whether the AOXs of gymnosperms, basal plants, eubacteria and animals are dimeric or monomeric. Examining the post-translational regulation of eubacterial and animal AOXs might identify a completely new mode of AOX regulation that is different from those of angiosperms, fungi and protists.
Genome-sequencing projects have allowed for the identification of AOX gene families in several plants and fungi, and when coupled with PCR and rapid amplification of cDNA ends technologies scientists can examine the 5′ and 3′ untranslated regions of AOX genes and their promoter regions in the hope of discovering how the transcription of AOX genes might be controlled (Chae et al. 2007a). Analyses of AOX promoters and the expression of AOX multigene families have revealed that each AOX gene is expressed differently and that gene orthology is not a reliable predictor of this information (Thirkettle-Watts et al. 2003). Further work in this area might aid us in determining how, when, and why multigene AOX families arose.
Microarray data and analyses of transcriptomes have identified transcripts that exhibit an expression pattern similar to one or more AOX genes. This has led to the discovery that an alternative mitochondrial NAD(P)H dehydrogenase is expressed in a similar pattern to AOX1a in A. thaliana, indicating that these two proteins might form a functional respiratory pathway together (Clifton et al. 2005). These newer technologies are making it possible to study the physiological function of AOX in novel ways.
Is there a single underlying function for AOX, or are there multiple functions that are as varied as the organisms that possess the enzyme? Was there a single ancestral function that has remained the same, or has it been successfully co-opted during evolution to fulfill new functions? It should be noted that the theories of the physiological function of AOX presented in this review are not necessarily mutually exclusive.
It is becoming increasingly clear that the moniker ‘alternative’ oxidase might have been somewhat premature given its apparent importance in the respiratory ETCs of a multitude of diverse organisms.
Acknowledgements
Research funding support by the Natural Sciences and Engineering Research Council of Canada and the Government of Ontario is gratefully acknowledged. I thank Dr Greg Vanlerberghe, Dr Norm Hüner and Dr Jim Staples for helpful discussions and constructive comments on drafts of the manuscript. I thank Dr Jennifer Watling and Dr Sharon Robinson for their early encouragement and support of this work. I thank the two anonymous reviewers for their constructive and insightful comments.
Abele E,
Philip E,
Gonzalez PM, Puntarulo S
(2007) Marine invertebrate mitochondria and oxidative stress. Frontiers in Bioscience 12, 933–946.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Affourtit C,
Albury MS,
Crichton PG, Moore AL
(2002) Exploring the molecular nature of alternative oxidase regulation and catalysis. FEBS Letters 510, 121–126.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Ajayi WU,
Chaudhuri M, Hill GC
(2002) Site-directed mutagenesis reveals the essentiality of the conserved residues in the putative diiron active site of the trypanosome alternative oxidase. Journal of Biological Chemistry 277, 8187–8193.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Akhter S,
McDade HC,
Gorlach JM,
Heinrich G,
Cox GM, Perfect JR
(2003) Role of alternative oxidase gene in pathogenesis of Cryptococcus neoformans. Infection and Immunity 71, 5794–5802.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Albury MS,
Affourtit C,
Crichton PG, Moore AL
(2002) Structure of the plant alternative oxidase. Site-directed mutagenesis provides new information on the active site and membrane topology. Journal of Biological Chemistry 277, 1190–1194.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Andersson ME, Nordlund P
(1999) A revised model of the active site of alternative oxidase. FEBS Letters 449, 17–22.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Angioy AM,
Stensmyr MC,
Urru I,
Puliafito M,
Collu I, Hansson BS
(2004) Function of the heater: the dead horse arum revisited. Proceedings. Biological Sciences/The Royal Society 271(Suppl.), S13–S15.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Archibald JM,
O’Kelly CJ, Doolittle WF
(2002) The chaperonin genes of jakobid and jakobid-like flagellates: implications for eukaryotic evolution. Molecular Biology and Evolution 19, 422–431.
| PubMed |

Arnholdt-Schmitt B,
Costa JH, de Melo DF
(2006) AOX—a functional marker for efficient cell reprogramming under stress? Trends in Plant Science 11, 281–287.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Atteia A,
van Lis R,
van Hellemond JJ,
Tielens AG,
Martin W, Henze K
(2004) Identification of prokaryotic homologues indicates an endosymbiotic origin for the alternative oxidases of mitochondria (AOX) and chloroplasts (PTOX). Gene 330, 143–148.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Azcón-Bieto J,
Ribas-Carbó M,
González-Meler M, Peñuelas J
(1989) Sulfide resistant respiration in leaves of Elodea canadensis Michx: comparison with cyanide-resistant respiration. Plant Physiology 90, 1249–1251.
| PubMed |

Baurain D,
Dinant M,
Coosemans N, Matagne RF
(2003) Regulation of the alternative oxidase Aox1 gene in Chlamydomonas reinhardtii. Role of the nitrogen source on the expression of a reporter gene under the control of the Aox1 promoter. Plant Physiology 131, 1418–1430.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Berthold DA
(1998) Isolation of mutants of the Arabidopsis thaliana alternative oxidase (ubiquinol : oxygen oxidoreductase) resistant to salicylhydroxamic acid. Biochimica et Biophysica Acta 1364, 73–83.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Berthold DA, Stenmark P
(2003) Membrane-bound diiron carboxylate proteins. Annual Review of Plant Biology 54, 497–517.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Berthold D,
Andersson ME, Nordlund P
(2000) New insight into the structure and function of the alternative oxidase. Biochimica et Biophysica Acta 1460, 241–254.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Berthold DA,
Voevodskaya N,
Graslund A, Nordlund P
(2002) EPR studies of the mitochondrial alternative oxidase. Evidence for a diiron carboxylate center. Journal of Biological Chemistry 277, 43608–43614.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Borecky J,
Nogueira FT,
de Oliveira KA,
Maia IG,
Vercesi AE, Arruda P
(2006) The plant energy-dissipating mitochondrial systems: depicting the genomic structure and the expression profiles of the gene families of uncoupling protein and alternative oxidase in monocots and dicots. Journal of Experimental Botany 57, 849–864.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Broadwater JA,
Ai J,
Loehr TM,
Sanders-Loehr J, Fox BG
(1998) Peroxodiferric intermediate of stearoyl-acyl carrier protein Δ9 desaturase: oxidase reactivity during single turnover and implications for the mechanism of desaturation. Biochemistry 37, 14664–14671.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Castro-Guerrero NA,
Krab K, Moreno-Sanchez R
(2004) The alternative respiratory pathway of Euglena mitochondria. Journal of Bioenergetics and Biomembranes 36, 459–469.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Castro-Guerrero NA,
Rodríguez-Zavala JS,
Marín-Hernández A,
Rodríguez-Enríquez S, Moreno-Sánchez R
(2008) Enhanced alternative oxidase and antioxidant enzymes under Cd2+ stress in Euglena. Journal of Bioenergetics and Biomembranes (In press). ,

Chae MS,
Lin CC,
Kessler KE,
Nargang CE,
Tanton LL,
Hahn LB, Nargang FE
(2007a) Identification of an alternative oxidase induction motif in the promoter region of the aod-1 gene in Neurospora crassa. Genetics 175, 1597–1606.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Chae MS,
Nargang CE,
Cleary IA,
Lin CC,
Todd AT, Nargang FE
(2007b) Two zinc-cluster transcription factors control induction of alternative oxidase in Neurospora crassa. Genetics 177, 1997–2006.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Chaudhuri M,
Sharan R, Hill GC
(2002) Trypanosome alternative oxidase is regulated post-transcriptionally at the level of RNA stability. Journal of Eukaryotic Microbiology 49, 263–269.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Chaudhuri M,
Ott RD,
Saha L,
Williams S, Hill GC
(2005) The trypanosome alternative oxidase exists as a monomer in Trypanosoma brucei mitochondria. Parasitology Research 96, 178–183.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Chaudhuri M,
Ott RD, Hill GC
(2006) Trypanosome alternative oxidase: from molecule to function. Trends in Parasitology 22, 484–491.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Claros MG, Vincens P
(1996) Computational method to predict mitochondrially imported proteins and their targeting sequences. European Journal of Biochemistry 241, 779–786.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Clifton R,
Lister R,
Parker KL,
Sappl PG,
Elhafez D,
Millar AH,
Day DA, Whelan J
(2005) Stress-induced co-expression of alternative respiratory chain components in Arabidopsis thaliana. Plant Molecular Biology 58, 193–212.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Clifton R,
Millar AH, Whelan J
(2006) Alternative oxidases in Arabidopsis: a comparative analysis of differential expression in the gene family provides new insights into function of non-phosphorylating bypasses. Biochimica et Biophysica Acta 1757, 730–741.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Considine MJ,
Daley DO, Whelan J
(2001) The expression of alternative oxidase and uncoupling protein during fruit ripening in mango. Plant Physiology 126, 1619–1629.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Considine MJ,
Holtzapffel RC,
Day DA,
Whelan J, Millar AH
(2002) Molecular distinction between alternative oxidase from monocots and dicots. Plant Physiology 129, 949–953.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Costa JH,
Jolivet Y,
Hasenfratz-Sauder MP,
Orellano EG,
da Guia Silva Lima M,
Dizengremel P, Fernandes de Melo D
(2007) Alternative oxidase regulation in roots of Vigna unguiculata cultivars differing in drought/salt tolerance. Journal of Plant Physiology 164, 718–727.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Cournac L,
Latouche G,
Cerovic Z,
Redding K,
Ravenel J, Peltier G
(2002) In vivo interactions between photosynthesis, mitorespiration, and chlororespiration in Chlamydomonas reinhardtii. Plant Physiology 129, 1921–1928.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Crichton PG,
Affourtit C,
Albury MS,
Carre JE, Moore AL
(2005) Constitutive activity of Sauromatum guttatum alternative oxidase in Schizosaccharomyces pombe implicates residues in addition to conserved cysteines in alpha-keto acid activation. FEBS Letters 579, 331–336.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Czarna M, Jarmuszkiewicz W
(2005) Activation of alternative oxidase and uncoupling protein lowers hydrogen peroxide formation in amoeba Acanthamoeba castellanii mitochondria. FEBS Letters 579, 3136–3140.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Czarna M,
Sluse FE, Jarmuszkiewicz W
(2007) Mitochondrial function plasticity in Acanthamoeba castellanii during growth in batch culture. Journal of Bioenergetics and Biomembranes 39, 149–157.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Day DA, Wiskich JT
(1995) Regulation of alternative oxidase in higher plants. Journal of Bioenergetics and Biomembranes 27, 379–385.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Day DA,
Krab K,
Lambers H,
Moore AL,
Siedow JN,
Wagner AM, Wiskich JT
(1996) The cyanide-resistant oxidase: to inhibit or not to inhibit, that is the question. Plant Physiology 110, 1–2.
| PubMed |

Deng Y,
Kohlwein SD, Mannella CA
(2002) Fasting induces cyanide-resistant respiration and oxidative stress in the amoeba Chaos carolinensis: implications for the cubic structural transition in mitochondrial membranes. Protoplasma 219, 160–167.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Descheneau AT,
Cleary IA, Nargang FE
(2005) Genetic evidence for a regulatory pathway controlling alternative oxidase production in Neurospora crassa. Genetics 169, 123–135.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Dinant M,
Baurain D,
Coosemans N,
Joris B, Matagne RF
(2001) Characterization of two genes encoding the mitochondrial alternative oxidase in Chlamydomonas reinhardtii. Current Genetics 39, 101–108.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Djajanegara I,
Holtzapffel R,
Finnegan PM,
Hoefnagel MH,
Berthold DA,
Wiskich JT, Day DA
(1999) A single amino acid change in the plant alternative oxidase alters the specificity of organic acid activation. FEBS Letters 454, 220–224.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Djajanegara I,
Finnegan PM,
Mathieu C,
McCabe T,
Whelan J, Day DA
(2002) Regulation of alternative oxidase gene expression in soybean. Plant Molecular Biology 50, 735–742.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Ederli L,
Morettini R,
Borgogni A,
Wasternack C,
Miersch O,
Reale L,
Ferranti F,
Tosti N, Pasqualini S
(2006) Interaction between nitric oxide and ethylene in the induction of alternative oxidase in ozone-treated tobacco plants. Plant Physiology 142, 595–608.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Elhafez D,
Murcha MW,
Clifton R,
Soole KL,
Day DA, Whelan J
(2006) Characterization of mitochondrial alternative NAD(P)H dehydrogenases in Arabidopsis: intraorganelle location and expression. Plant & Cell Physiology 47, 43–54.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Fang J, Beattie DS
(2003) Alternative oxidase present in procyclic Trypanosoma brucei may act to lower the mitochondrial production of superoxide. Archives of Biochemistry and Biophysics 414, 294–302.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Finnegan PM,
Umbach AL, Wilce JA
(2003) Prokaryotic origins for the mitochondrial alternative oxidase and plastid terminal oxidase nuclear genes. FEBS Letters 555, 425–430.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Fisher N, Rich PR
(2000) A motif for quinone binding sites in respiratory and photosynthetic systems. Journal of Molecular Biology 296, 1153–1162.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Fung RW,
Wang CY,
Smith DL,
Gross KC,
Tao Y, Tian M
(2006) Characterization of alternative oxidase (AOX) gene expression in response to methyl salicylate and methyl jasmonate pre-treatment and low temperature in tomatoes. Journal of Plant Physiology 163, 1049–1060.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Gassner GT, Lippard SJ
(1999) Component interactions in the soluble methane monooxygenase system from Methylococcus capsulatus (Bath). Biochemistry 38, 12768–12785.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Gelhaye E,
Rouhier N,
Gerard J,
Jolivet Y, Gualberto J , et al..
(2004) A specific form of thioredoxin h occurs in plant mitochondria and regulates the alternative oxidase. Proceedings of the National Academy of Sciences of the United States of America 101, 14545–14550.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Gomes CM,
Le Gall J,
Xavier AV, Teixeira M
(2001) Could a diiron-containing four-helix-bundle protein have been a primitive oxygen reductase? ChemBioChem: A European Journal of Chemical Biology 2, 583–587.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Gray GR,
Villarimo AR,
Whitehead CL, McIntosh L
(2004) Transgenic tobacco (Nicotiana tabacum L.) plants with increased expression levels of mitochondrial NADP+-dependent isocitrate dehydrogenase: evidence implicating this enzyme in the redox activation of the alternative oxidase. Plant & Cell Physiology 45, 1413–1425.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Gredilla R,
Grief J, Osiewacz HD
(2006) Mitochondrial free radical generation and lifespan control in the fungal aging model Podospora anserina. Experimental Gerontology 41, 439–447.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Grieshaber MK, Völkel S
(1998) Animal adaptations for tolerance and exploitation of poisonous sulfide. Annual Review of Physiology 60, 33–53.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Guy RD,
Berry JA,
Fogel ML, Hoering TC
(1989) Differential fractionation of oxygen isotopes by cyanide-resistant and cyanide-sensitive respiration in plants. Planta 177, 483–491.
| Crossref | GoogleScholarGoogle Scholar |

Hakkaart GA,
Dassa EP,
Jacobs HT, Rustin P
(2006) Allotopic expression of a mitochondrial alternative oxidase confers cyanide resistance to human cell respiration. EMBO Reports 7, 341–345.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Hanssens L, Verachtert H
(1976) Adenosine 5′-monophosphate-stimulated cyanide-insensitive respiration in mitochondria of Moniliella tomentosa. Journal of Bacteriology 125, 829–836.

Henry M-F, Nyns E-J
(1975) Cyanide-insensitive respiration. An alternative mitochondrial pathway. Sub-Cellular Biochemistry 4, 1–65.
| PubMed |

Ho LH,
Giraud E,
Lister R,
Thirkettle-Watts D,
Low J,
Clifton R,
Howell KA,
Carrie C,
Donald T, Whelan J
(2007) Characterization of the regulatory and expression context of an alternative oxidase gene provides insights into cyanide-insensitive respiration during growth and development. Plant Physiology 143, 1519–1533.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Hoefnagel MHN, Wiskich JT
(1998) Activation of the plant alternative oxidase by high reduction levels of the Q-pool and pyruvate. Archives of Biochemistry and Biophysics 355, 262–270.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Huang X,
von Rad U, Durner J
(2002) Nitric oxide induces transcriptional activation of the nitric oxide-tolerant alternative oxidase in Arabidopsis suspension cells. Planta 215, 914–923.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Huh WK, Kang SO
(2001) Characterization of the gene family encoding alternative oxidase from Candida albicans. The Biochemical Journal 356, 595–604.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Ito Y,
Saisho D,
Nakazono M,
Tsutsumi N, Hirai A
(1997) Transcript levels of tandem-arranged alternative oxidase genes in rice are increased by low temperature. Gene 203, 121–129.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Jarmuszkiewicz W,
Wagner AM,
Wagner MJ, Hryniewiecka L
(1997) Immunological identification of the alternative oxidase of Acanthamoeba castellanii mitochondria. FEBS Letters 411, 110–114.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Jarmuszkiewicz W,
Behrendt M,
Navet R, Sluse FE
(2002) Uncoupling protein and alternative oxidase of Dictyostelium discoideum: occurrence, properties and protein expression during vegetative life and starvation-induced early development. FEBS Letters 532, 459–464.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Jarmuszkiewicz W,
Czarna M, Sluse FE
(2005) Substrate kinetics of the Acanthamoeba castellanii alternative oxidase and the effects of GMP. Biochimica et Biophysica Acta 1708, 71–78.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Joseph-Horne T,
Babij J,
Wood PM,
Hollomon D, Sessions RB
(2000) New sequence data enable modelling of the fungal alternative oxidase and explain an absence of regulation by pyruvate. FEBS Letters 481, 141–146.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Juárez O,
Guerra G,
Martinez F, Pardo JP
(2004) The mitochondrial respiratory chain of Ustilago maydis. Biochimica et Biophysica Acta 1658, 244–251.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Juárez O,
Guerra G,
Velázquez I,
Flores-Herrera O,
Rivera-Pérez RE, Pardo JP
(2006) The physiologic role of alternative oxidase in Ustilago maydis. The FEBS Journal 273, 4603–4615.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Juszczuk IM, Rychter AM
(2003) Alternative oxidase in higher plants. Acta Biochimica Polonica 50, 1257–1271.
| PubMed |

Karpova OV,
Kuzmin EV,
Elthon TE, Newton KJ
(2002) Differential expression of alternative oxidase genes in maize mitochondrial mutants. The Plant Cell 14, 3271–3284.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Kirimura K,
Ogawa S,
Hattori T, Kino K
(2006) Expression analysis of alternative oxidase gene (aox1) with enhanced green fluorescent protein as marker in citric acid-producing Aspergillus niger. Journal of Bioscience and Bioengineering 102, 210–214.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Laloi C,
Rayapuram N,
Chartier Y,
Grienenberger J-M,
Bonnard G, Meyer Y
(2001) Identification and characterization of a mitochondrial thioredoxin system in plants. Proceedings of the National Academy of Sciences of the United States of America 98, 14144–14149.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Lambers H
(1982) Cyanide resistant respiration: a non-phosphorylating electron transport pathway acting as an overflow. Physiologia Plantarum 55, 478–485.
| Crossref | GoogleScholarGoogle Scholar |

Lang BF,
O’Kelly C,
Nerad T,
Gray MW, Burger G
(2002) The closest unicellular relatives of animals. Current Biology 12, 1773–1778.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Li F,
Zhang Y,
Wang M,
Zhang Y,
Wu X, Guo X
(2008) Molecular cloning and expression characteristics of alternative oxidase gene of cotton (Gossypium hirsutum). Molecular Biology Reports 35, 97–105.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Li Q,
Ritzel RG,
McLean LL,
McIntosh L,
Ko T,
Bertrand H, Nargang FE
(1996) Cloning and analysis of the alternative oxidase gene of Neurospora crassa. Genetics 142, 129–140.
| PubMed |

Magnani T,
Soriani FM,
Martins VP,
Nascimento AM,
Tudella VG,
Curti C, Uyemura SA
(2007) Cloning and functional expression of the mitochondrial alternative oxidase of Aspergillus fumigatus and its induction by oxidative stress. FEMS Microbiology Letters 271, 230–238.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Mathy G,
Navet R,
Gerkens P,
Leprince P,
De Pauw E,
Sluse-Goffart CM,
Sluse FE, Douette P
(2006) Saccharomyces cerevisiae mitoproteome plasticity in response to recombinant alternative ubiquinol oxidase. Journal of Proteome Research 5, 339–348.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Maxwell DP,
Wang Y, McIntosh L
(1999) The alternative oxidase lowers mitochondrial reactive oxygen production in plant cells. Proceedings of the National Academy of Sciences of the United States of America 96, 8271–8276.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

McDonald AE, Vanlerberghe GC
(2004) Branched mitochondrial electron transport in the Animalia: presence of alternative oxidase in several animal phyla. IUBMB Life 56, 333–341.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

McDonald AE, Vanlerberghe GC
(2005) Alternative oxidase and plastoquinol terminal oxidase in marine prokaryotes of the Sargasso Sea. Gene 349, 15–24.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

McDonald AE, Vanlerberghe GC
(2006) Origins, evolutionary history, and taxonomic distribution of alternative oxidase and plastoquinol terminal oxidase. Comparative Biochemistry and Physiology. Part D, Genomics & Proteomics 1, 357–364.
| Crossref | GoogleScholarGoogle Scholar |

McDonald AE,
Amirsadeghi S, Vanlerberghe GC
(2003) Prokaryotic orthologues of mitochondrial alternative oxidase and plastid terminal oxidase. Plant Molecular Biology 53, 865–876.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Medentsev AG,
Arinbasarova AI,
Smirnova NM, Akimenko VK
(2004) Activation of the alternative oxidase of Yarrowia lipolytica by adenosine 5′-monophosphate. Mikrobiologiia 73, 149–156.
| PubMed |

Millar AH,
Wiskich JT,
Whelan J, Day DA
(1993) Organic acid activation of the alternative oxidase of plant mitochondria. FEBS Letters 329, 259–262.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Millenaar FF, Lambers H
(2003) The alternative oxidase: in vivo regulation and function. Plant Biology 5, 2–15.
| Crossref | GoogleScholarGoogle Scholar |

Møller IM
(2001) Plant mitochondria and oxidative stress: electron transport, NADPH turnover, and metabolism of reactive oxygen species. Annual Review of Plant Physiology 52, 561–591.
| Crossref | GoogleScholarGoogle Scholar |

Møller IM,
Berczi A,
van der Plas LHW, Lambers H
(1988) Measurement of the activity and capacity of the alternative pathway in intact plant tissues: identification of problems and possible solutions. Physiologia Plantarum 72, 642–649.
| Crossref | GoogleScholarGoogle Scholar |

Moore AL,
Umbach AL, Siedow JN
(1995) Structure function relationships of the alternative oxidase of plant mitochondria—a model of the active site. Journal of Bioenergetics and Biomembranes 27, 367–377.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Moore AL,
Carré JE,
Affourtit C,
Albury MS,
Crichton PG,
Kita K, Heathcote P
(2008) Compelling EPR evidence that the alternative oxidase is a diiron carboxylate protein. Biochimica et Biophysica Acta 1777, 327–330.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Nakamura K,
Sakamoto K,
Kido Y,
Fujimoto Y, Suzuki T , et al..
(2005) Mutational analysis of the Trypanosoma vivax alternative oxidase: the E(X)6Y motif is conserved in both mitochondrial alternative oxidase and plastid terminal oxidase and is indispensable for enzyme activity. Biochemical and Biophysical Research Communications 334, 593–600.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Nicholls P
(1975) The effect of sulfide on cytochrome aa3. Isosteric and allosteric shifts of the reduced a-peak. Biochimica et Biophysica Acta 396, 24–35.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Nihei C,
Fukai Y,
Kawai K,
Osanai A,
Yabu Y,
Suzuki T,
Ohta N,
Minagawa N,
Nagai K, Kita K
(2003) Purification of active recombinant trypanosome alternative oxidase. FEBS Letters 538, 35–40.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Noguchi K, Yoshida K
(2008) Interaction between photosynthesis and respiration in illuminated leaves. Mitochondrion 8, 87–99.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Onda Y,
Kato Y,
Abe Y,
Ito T, Ito-Inaba Y , et al..
(2007) Pyruvate-sensitive AOX exists as a non-covalently associated dimer in the homeothermic spadix of the skunk cabbage, Symplocarpus renifolius. FEBS Letters 581, 5852–5858.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Ott R,
Chibale K,
Anderson S,
Chipeleme A,
Chaudhuri M,
Guerrah A,
Colowick N, Hill GC
(2006) Novel inhibitors of the trypanosome alternative oxidase inhibit Trypanosoma brucei brucei growth and respiration. Acta Tropica 100, 172–184.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Pastore D,
Trono D,
Laus MN,
Di Fonzo N, Passarella S
(2001) Alternative oxidase in durum wheat mitochondria. Activation by pyruvate, hydroxypyruvate and glyoxylate and physiological role. Plant & Cell Physiology 42, 1373–1382.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Pastore D,
Trono D,
Laus MN,
Di Fonzo N, Flagella Z
(2007) Possible plant mitochondria involvement in cell adaptation to drought stress. A case study: durum wheat mitochondria. Journal of Experimental Botany 58, 195–210.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Polidoros AN,
Mylona PV,
Pasentsis K,
Scandalios JG, Tsaftaris AS
(2005) The maize alternative oxidase 1a (Aox1a) gene is regulated by signals related to oxidative stress. Redox Report 10, 71–78.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Popov VN,
Purvis AC,
Skulachev VP, Wagner AM
(2001) Stress-induced changes in ubiquinone concentration and alternative oxidase in plant mitochondria. Bioscience Reports 21, 369–379.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Putignani L,
Tait A,
Smith HV,
Horner D,
Tovar J,
Tetley L, Wastling JM
(2004) Characterization of a mitochondrion-like organelle in Cryptosporidium parvum. Parasitology 129, 1–18.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Reyes-Prieto A,
El-Hafidi M,
Moreno-Sanchez R, Gonzalez-Halphen D
(2002) Characterization of oxidative phosphorylation in the colorless chlorophyte Polytomella sp. Its mitochondrial respiratory chain lacks a plant-like alternative oxidase. Biochimica et Biophysica Acta 1554, 170–179.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Rhoads DM, McIntosh L
(1993) The salicylic acid-inducible alternative oxidase gene aox1 and genes encoding pathogenesis-related proteins share regions of sequence similarity in their promoters. Plant Molecular Biology 21, 615–624.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Rhoads DM, Subbaiah CC
(2007) Mitochondrial retrograde regulation in plants. Mitochondrion 7, 177–194.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Rhoads DM,
Umbach AL,
Sweet CR,
Lennon AM,
Rauch GS, Siedow JN
(1998) Regulation of the cyanide-resistant alternative oxidase of plant mitochondria. Identification of the cysteine residue involved in alpha-keto acid stimulation and intersubunit disulfide bond formation. Journal of Biological Chemistry 273, 30750–30756.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Roberts CW,
Roberts F,
Henriquez FL,
Akiyoshi D, Samuel BU , et al..
(2004) Evidence for mitochondrial-derived alternative oxidase in the apicomplexan parasite Cryptosporidium parvum: a potential anti-microbial agent target. International Journal for Parasitology 34, 297–308.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Robinson SA,
Ribas-Carbo M,
Yakir D,
Giles L,
Reuveni Y, Berry JA
(1995) Beyond SHAM and cyanide: opportunities for studying the alternative oxidase in plant respiration using oxygen isotope discrimination. Functional Plant Biology 22, 487–496.
| Crossref | GoogleScholarGoogle Scholar |

Saika H,
Ohtsu K,
Hamanaka S,
Nakazono M,
Tsutsumi N, Hirai A
(2002) AOX1c, a novel rice gene for alternative oxidase: comparison with rice AOX1a and AOX1b. Genes & Genetic Systems 77, 31–38.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Saisho D,
Nambara E,
Naito S,
Tsutsumi N,
Hirai A, Nakazono M
(1997) Characterization of the gene family for alternative oxidase from Arabidopsis thaliana. Plant Molecular Biology 35, 585–596.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Sazinsky MH, Lippard SJ
(2006) Correlating structure with function in bacterial multicomponent monooxygenases and related diiron proteins. Accounts of Chemical Research 39, 558–566.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Schloss PD, Hendelsman J
(2005) Metagenomics for studying unculturable microorganisms: cutting the Gordian knot. Genome Biology 6, 229–232.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Siedow JN,
Umbach AL, Moore AL
(1995) The active site of the cyanide-resistant oxidase from plant mitochondria contains a binuclear iron center. FEBS Letters 362, 10–14.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Stenmark P, Nordlund P
(2003) A prokaryotic alternative oxidase present in the bacterium Novosphingobium aromaticivorans. FEBS Letters 552, 189–192.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Suzuki T,
Hashimoto T,
Yabu Y,
Kido Y, Sakamoto K , et al..
(2004) Direct evidence for cyanide-insensitive quinol oxidase (alternative oxidase) in the apicomplexan parasite Cryptosporidium parvum: phylogenetic and therapeutic implications. Biochemical and Biophysical Research Communications 313, 1044–1052.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Takumi S,
Tomioka M,
Eto K,
Naydenov N, Nakamura C
(2002) Characterization of two non-homoeologous nuclear genes encoding mitochondrial alternative oxidase in common wheat. Genes & Genetic Systems 77, 81–88.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Tanudji M,
Sjoling S,
Glaser E, Whelan J
(1999) Signals required for the import and processing of the alternative oxidase into mitochondria. Journal of Biological Chemistry 274, 1286–1293.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Theissen U,
Hoffmeister M,
Grieshaber M, Martin W
(2003) Single eubacterial origin of eukaryotic sulfide : quinone oxidoreductase, a mitochondrial enzyme conserved from the early evolution of eukaryotes during anoxic and sulfidic times. Molecular Biology and Evolution 20, 1564–1574.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Thirkettle-Watts D,
McCabe TC,
Clifton R,
Moore C,
Finnegan PM,
Day DA, Whelan J
(2003) Analysis of the alternative oxidase promoters from soybean. Plant Physiology 133, 1158–1169.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Thompson JD,
Gibson TJ,
Plewniak F,
Jeanmougin F, Higgins DG
(1997) The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research 25, 4876–4882.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Tringe SG, Rubin EM
(2005) Metagenomics: DNA sequencing of environmental samples. Nature Reviews. Genetics 6, 805–814.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Umbach AL, Siedow JN
(1993) Covalent and noncovalent dimers of the cyanide-resistant alternative oxidase protein in higher plant mitochondria and their relationship to enzyme activity. Plant Physiology 103, 845–854.
| PubMed |

Umbach AL, Siedow JN
(1996) The reaction of the soybean cotyledon mitochondrial cyanide-resistant oxidase with sulfhydryl reagents suggests that α-keto acid activation involves the formation of a thiohemiacetal. Journal of Biological Chemistry 271, 25019–25026.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Umbach AL, Siedow JN
(2000) The cyanide-resistant alternative oxidases from the fungi Pichia stipitis and Neurospora crassa are monomeric and lack regulatory features of the plant enzyme. Archives of Biochemistry and Biophysics 378, 234–245.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Umbach AL,
Wiskich JT, Siedow JN
(1994) Regulation of alternative oxidase kinetics by pyruvate and intermolecular disulfide bond redox status in soybean seedling mitochondria. FEBS Letters 348, 181–184.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Umbach AL,
Gonzàlez-Meler MA,
Sweet CR, Siedow JN
(2002) Activation of the plant mitochondrial alternative oxidase: insights from site-directed mutagenesis. Biochimica et Biophysica Acta 1554, 118–128.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Van Hellemond JJ,
Simons B,
Millenaar FF, Tielens AG
(1998) A gene encoding the plant-like alternative oxidase is present in Phytomonas but absent in Leishmania spp. Journal of Eukaryotic Microbiology 45, 426–430.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Vanderleyden J,
Peeters C,
Verachtert H, Bertrand H
(1980) Stimulation of the alternative oxidase of Neurospora crassa by nucleoside phosphates. The Biochemical Journal 188, 141–144.
| PubMed |

Vanlerberghe GC, McIntosh L
(1996) Signals regulating the expression of the nuclear gene encoding alternative oxidase of plant mitochondria. Plant Physiology 111, 589–595.
| PubMed |

Vanlerberghe GC,
Vanlerberghe AE, McIntosh L
(1994) Molecular genetic alteration of plant respiration (silencing and overexpression of alternative oxidase in transgenic tobacco). Plant Physiology 106, 1503–1510.
| PubMed |

Vanlerberghe G,
Day D,
Wiskich JT,
Vanlerberghe AE, McIntosh L
(1995) Alternative oxidase activity in tobacco leaf mitochondria: dependence on tricarboxylic acid cycle-mediated redox regulation and pyruvate activation. Plant Physiology 109, 353–361.
| PubMed |

Vanlerberghe GC,
Vanlerberghe AE, McIntosh L
(1997) Molecular genetic evidence of the ability of alternative oxidase to support respiratory carbon metabolism. Plant Physiology 113, 657–661.
| PubMed |

Vanlerberghe GC,
McIntosh L, Yip JYH
(1998) Molecular localization of a redox-modulated process regulating plant mitochondrial electron transport. The Plant Cell 10, 1551–1560.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Vanlerberghe GC,
Yip JY, Parsons HL
(1999) In organello and in vivo evidence of the importance of the regulatory sulfhydryl/disulfide system and pyruvate for alternative oxidase activity in tobacco. Plant Physiology 121, 793–803.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Veiga A,
Arrabaca JD, Loureiro-Dias MC
(2003) Cyanide-resistant respiration, a very frequent metabolic pathway in yeasts. FEMS Yeast Research 3, 239–245.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Venter JC,
Remington K,
Heidelberg JF,
Halpern AL, Rusch D , et al..
(2004) Environmental genome shotgun sequencing of the Sargasso Sea. Science 304, 66–74.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Watling JR,
Robinson SA, Seymour RS
(2006) Contribution of the alternative pathway to respiration during thermogenesis in flowers of the sacred lotus. Plant Physiology 140, 1367–1373.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Whelan J,
Smith MK,
Meijer M,
Yu JW,
Badger MR,
Price GD, Day DA
(1995) Cloning of an additional cDNA for the alternative oxidase in tobacco. Plant Physiology 107, 1469–1470.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Whelan J,
Millar AH, Day DA
(1996) The alternative oxidase is encoded in a multigene family in soybean. Planta 198, 197–201.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Yip JY, Vanlerberghe GC
(2001) Mitochondrial alternative oxidase acts to dampen the generation of active oxygen species during a period of rapid respiration induced to support a high rate of nutrient uptake. Physiologia Plantarum 112, 327–333.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Yoshida K,
Terashima I, Noguchi K
(2007) Up-regulation of mitochondrial alternative oxidase concomitant with chloroplast over-reduction by excess light. Plant & Cell Physiology 48, 606–614.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Yukioka H,
Inagaki S,
Tanaka R,
Katoh K,
Miki N,
Mizutani A, Masuko M
(1998) Transcriptional activation of the alternative oxidase gene of the fungus Magnaporthe grisea by a respiratory-inhibiting fungicide and hydrogen peroxide. Biochimica et Biophysica Acta 1442, 161–169.
| PubMed |