Analysis of global host gene expression during the primary phase of the Arabidopsis thaliana–Plasmodiophora brassicae interaction
Arati Agarwal A B F , Vijay Kaul B C , Robert Faggian D , James E. Rookes A , Jutta Ludwig-Müller E and David M. Cahill AA School of Life and Environmental Sciences, Deakin University, Geelong Campus at Waurn Ponds, Vic. 3217, Australia.
B Department of Primary Industries, Private Bag 15, Ferntree Gully DC, Vic. 3156, Australia.
C Present address: School of Botany, The University of Melbourne, Parkville, Vic. 3010, Australia.
D Department of Primary Industries, 32 Lincoln Square Nth Carlton, PO Box 4166, Parkville, Vic. 3052, Australia.
E Department of Biology, Technische Universität Dresden, 01062 Dresden, Germany.
F Corresponding author. Email: arati.agarwal@dpi.vic.gov.au
Functional Plant Biology 38(6) 462-478 https://doi.org/10.1071/FP11026
Submitted: 24 January 2011 Accepted: 31 March 2011 Published: 3 June 2011
Abstract
Microarray analysis was used to investigate changes in host gene expression during the primary stages of the interaction between the susceptible plant Arabidopsis thaliana (L.) Heynh ecotype Col-0 and the biotrophic pathogen Plasmodiophora brassicae Woronin. Analyses were conducted at 4, 7 and 10 days after inoculation (DAI) and revealed significant induction or suppression of a relatively low number of genes in a range of functional categories. At 4 DAI, there was induced expression of several genes known to be critical for pathogen recognition and signal transduction in other resistant host–pathogen interactions. As the pathogen further colonised root tissue and progressed through the primary plasmodium stage to production of zoosporangia at 7 and 10 DAI, respectively, fewer genes showed changes in expression. The microarray results were validated by examining a subset of induced genes at 4 DAI by quantitative real-time reverse transcriptase PCR (RT-qPCR) analysis all of which correlated positively with the microarray data. The two A. thaliana mutants jar1 and coiI tested were found to be susceptible to P. brassicae. The involvement of defence-related hormones in the interaction was further investigated and the findings indicate that addition of salicylic acid can suppress clubroot disease in A. thaliana plants.
Additional keywords: ATH1 microarray chip, clubroot, compatibility, real-time RT-qPCR, salicylic acid.
Introduction
Plasmodiophora brassicae Woronin is an obligate biotrophic root pathogen that attacks brassicaceous plant species and can cause significant yield losses (Braselton 1995; Dixon 2009). Control strategies are limited to the use of crop rotation, liming, and application of calcium and boron in many parts of the world (Donald and Porter 2009). Similarly, breeding for resistant cultivars has been problematic in brassica crops due to the wide variation in virulence of field populations of P. brassicae (Crute et al. 1980; Toxopeus et al. 1986; Hirai 2006). Wide-scale gene expression analysis offers an opportunity to explore the molecular basis of plant–pathogen interactions, particularly with respect to mechanisms of resistance and basal defence, as well as host versus non-host resistance, and biotrophy versus necrotrophy (Oliver and Ipcho 2004; Anderson et al. 2005; Glazebrook 2005; Wise et al. 2007). Microarray technology has enabled the identification of defence-related genes for many plant–pathogen interactions as well as the mechanisms by which plants develop resistance. In addition, several studies show that biotrophs have the ability to evade recognition and suppress defence activation in order to establish compatibility in the host plant (Schulze-Lefert and Panstruga 2003; da Cunha et al. 2007). There is limited knowledge at the molecular and genetic level of host compatibility and how virulent pathogens may evade detection, suppress host defences or both.
The lifecycle of P. brassicae occurs in two distinct phases: the primary phase, which occurs predominantly in root hairs, and the secondary phase, which occurs predominantly in the root cortex (Ingram and Tommerup 1972). It is the secondary phase that produces the characteristic disease symptoms of root tissue hypertrophy and hyperplasia, or ‘clubbing’. Broadly speaking, in resistant hosts, the primary phase of the lifecycle can still be observed, but not the secondary phase (Dekhuijzen 1979; Takahashi et al. 2006; Donald et al. 2008). In a compatible (susceptible) interaction with Arabidopsis thaliana (L.) Heynh., the pathogen can proceed through its entire lifecycle from 3 to 28 days, as studied by Siemens et al. (2002). Recent work has demonstrated that P. brassicae attachment to, and penetration of, the root hair can occur from Day 4 after inoculation onwards (Agarwal et al. 2009). This occurs after a resting spore releases a single motile zoospore that attaches to the wall of the root hair and encysts by means of a tubular structure (the rohr and the stachel), injecting its protoplast into the host cytoplasm (Aist and Williams 1971). Between 5 and 12 days after inoculation, plasmodia and then zoosporangia (producing secondary zoospores) develop within the root hairs with little visual sign of damage to the host cells. Between 10 and 13 days after inoculation, P. brassicae undergoes a transition from the primary phase to the secondary phase of its lifecycle as observed by Devos et al. (2005) and Siemens et al. (2006) depending on the isolate used (single spore or field population). Within 28 days after inoculation, the secondary plasmodia multiply into resting spores within the cortical cells proliferating into galls in A. thaliana (Agarwal et al. 2009).
Siemens et al. (2006) have reported the use of microarray analysis to examine gene expression following inoculation of a susceptible A. thaliana ecotype Col-0 with P. brassicae at 10 and 23 days after inoculation (DAI) using a single spore isolate. These two time points were within the so-called secondary infection stage, which takes place in the root cortex – the 10-day time point being described as the early secondary plasmodial stage and the 23-day time point as the late secondary stage, by which time the disease has progressed substantially in the roots to develop galls. Their study found wide-scale changes in the expression of genes in several important functional groups including those involved in growth, sugar phosphate metabolism, defence and plant hormone synthesis. The two time points analysed, however, were relatively late in the interaction for studying the potentially important events that take place during initial pathogen invasion. Proteomic-based approaches have also been reported (Devos et al. 2006; Cao et al. 2008). Devos et al. (2006) examined the susceptible response in A. thaliana roots at 4 DAI with a field population of P. brassicae. Most of the differentially regulated proteins (12%) were involved in plant defence, cell differentiation, hormone metabolism and detoxification. More recently, Cao et al. (2008) examined the susceptible response in canola (Brassica napus L.) roots 12, 24, 48 and 72 h after inoculation using the field population of P. brassicae collected from central Alberta, Canada. Only 20 protein spots in total were differentially regulated including proteins involved in lignin biosynthesis, cytokinin metabolism, glycolysis, intracellular calcium homeostasis and the detoxification of reactive oxygen species (ROS). Furthermore, a review of the interactions of P. brassicae with its hosts (Ludwig-Müller and Schuller 2008) once again emphasised that although the components of early signalling in the interaction between A. thaliana and leaf pathogens are well understood, there are relatively few studies with root pathogens and indeed none conducted with P. brassicae. A comparative study between resistance and susceptibility is not possible for the interaction of P. brassicae with A. thaliana, as almost all the A. thaliana ecotypes tested so far are susceptible to P. brassicae (Siemens et al. 2002; Agarwal et al. 2009). Also, the enormous variation in the virulence of P. brassicae populations reported worldwide (Jones et al. 1982) often makes it difficult to determine the host’s genomic and metabolomic responses.
This research reports wide-scale gene expression analysis for this host–pathogen interaction up to 10 DAI, when the pathogen is still in the primary phase of its lifecycle. This study examined the early critical time points (4, 7 and 10 DAI) of the susceptible response of the A. thaliana ecotype Col-0 following inoculation with a field population of P. brassicae. Changes in gene expression were analysed using the Arabidopsis ATH1 GeneChip array (Affymetrix, Santa Clara, CA, USA) to explore the basis of this compatible interaction.
Materials and methods
Inoculum preparation, plant growth, salicylic acid treatment and disease assessment
Plasmodiophora brassicae Woronin inoculum was prepared from mature galls on Brassica oleracea L. (broccoli) roots European Clubroot Differential (ECD) code (pathotype) 16/19/31 collected from a vegetable farm in Werribee South, Vic., Australia. Following collection, root material containing galls were stored at −20°C (Department of Primary Industries, Knoxfield, Vic., Australia). Galls taken from frozen roots were used for extraction of resting spores by homogenisation in deionised water (1 : 3) (w : v) followed by filtration of the crude extract through a double gauze filter (25 µm pore width). Spore density was determined using a haemocytometer (Neubauer Improved, Germany) and suspensions of 108 spores mL–1 in water were prepared for inoculation.
Arabidopsis thaliana (L.) Heynh. plants (ecotype Col-0) were grown within pots in seed raising mix (Debco, Tyabb, Vic., Australia). The seeds were first stratified at 4°C in the dark for 4 days and then pots 120 mm in diameter were transferred to a growth room with controlled environment conditions at 20°C, with 75% relative humidity and a 16-h photoperiod at 100 µmol m–2 s–1. Once seedlings reached 14 days of age, they were singled out in small pots 60 mm in diameter, and 20 pots were arranged randomly within one tray. Each pot was inoculated with 200 μL of a P. brassicae suspension (108 spores mL–1) by pipetting in and around the soil adjacent to the plant roots; the pots were then transferred back to the growth room. Control plants were mock-inoculated with water. Three independent experiments were conducted to produce the dataset. Plants were uprooted at 0, 4, 7 and 10 DAI from both challenged and unchallenged plants. Soil particles were removed from roots by gently washing in water. As preliminary experiments had shown that a larger number of root systems were required at day 0 and day 4 because of the small root size, roots from 50 plants at Days 0 and 4 and 20 plants at Days 7 and 10 were collected. Roots of plants from the same time point were pooled together, kept in 1.5-mL Eppendorf tubes (Axygen, Union City, CA, USA), followed by freezing tubes in liquid nitrogen and then stored at −80°C for RNA extraction. Twenty-five to thirty plants within each replicate were left to grow on until 28 DAI for the purpose of disease assessment. A. thaliana plants were carefully uprooted from the soil at intervals up to a maximum of 28 DAI and observed for macroscopic symptoms of infection induced in both roots and shoots.
In a separate experiment, 14-day-old A. thaliana seedlings grown under the same conditions as mentioned above were carefully removed from the pots. The roots were gently washed and dipped in 500 µM unbuffered salicylic acid (BDH Chemicals, Poole, England) for 60 s before being transferred back to soil drenched with water in individual pots (20 pots arranged randomly within one tray). The inoculum was prepared as mentioned above. After 4 h equilibration in the growth chamber, plants were either (i) inoculated with a 200 µL suspension of P. brassicae resting spores (108 spores mL–1), or (ii) mock-inoculated with 200 µL of sterile distilled water. Another subset of control plants, which were not treated with salicylic acid, was also inoculated with P. brassicae as described above.
The plants were gently removed from soil 50 DAI and the roots were then washed with water and visually assessed. The disease index (DI) for A. thaliana was calculated based on a scale consisting of five classes (0–4) according to published protocols (Kobelt et al. 2000) where: 0 = no symptoms; 1 = very small clubs, mainly on lateral roots, which do not impair the main root; 2 = small clubs covering the main root and a few lateral roots; 3 = medium-sized to bigger clubs, also including the main root (up to two-thirds), possibly impairing plant growth; and 4 = severe clubs in lateral roots and main root, fine roots completely destroyed, and plant growth also affected. The DI was calculated using the five-grade scale according to the formula: DI = (0n0 + 1n1 + 2n2 + 3n3 + 4n4) × 100/4Nt, where n0 to n4 is the number of plants in the indicated class and Nt is the total number of plants tested. Depending on the DI calculated for each host, the plant was classified as resistant (DI = 0) or susceptible (DI ≥ 30). A DI of 100 = full susceptibility and DI > 0 < 30 = partial resistance (restriction of pathogen growth in host) or tolerance (no restriction of pathogen but no yield loss). Infected plants (IP) were expressed as the percentage of the total number of plants infected by the total number of plants tested.
RNA extraction
Total RNA was extracted using the RNAqueous kit (Ambion, Austin, TX, USA) for samples from each of the three replicates. Each RNA sample was further purified to remove contaminants such as polysaccharides and polyphenolics (RNA Isolation Aid, Ambion). Lithium chloride (Ambion) precipitation was further performed to enhance the purity of the RNA sample by removing carbohydrates and gross DNA contamination. RNA was quantified using spectrophotometry (NanoDrop Technologies, Wilmington, DE, USA) and stored at −20°C. The RNA quality was assessed by running 1 μL of each sample on a RNA Laboratory-On-A-Chip (Agilent Bioanalyser 2100 Agilent Technologies, Palo Alto, CA, USA).
Target preparation, synthesis and microarray hybridisation
For each test sample, 100 ng of total RNA was labelled for expression analysis according to the Two-Cycle Eukaryotic Target Labelling Assay (Affymetrix). RNA target preparation, target hybridisation, and washing, staining and scanning of arrays were performed according to the protocol described in the GeneChip Expression Analysis Technical Manual (Affymetrix). A software package (GeneChip Operating System (GCOS) version 1.2 (Affymetrix)) was used for image analysis, for checking probe signal levels, and for generating and analysing the data. The first step was to perform ‘absolute analysis’ to assess the hybridisation between targets (cRNA derived from mRNA) and probes. A scaling factor of 150 was used for normalising each probe array for converting the fluorescence intensity of probe cell to a signal value. Based on probe pair intensities, a detection P-value was calculated that indicated the probability of a detection call being correct. Absolute analysis generated signal values for all 22 810 probe sets on the ATH1 array. The quality of every array’s scanned image was checked for noise, background, the percentage of genes present and absent, housekeeping genes and spiked controls. The reports generated for all arrays were comparable, suggesting a high level of reproducibility. More than 55% of the probes called present in all 21 chips exhibited a variation of less than 10%.
Microarray data analysis and data collection
The software package AVADIS (Accessing, Visualising, Analysing and Discover version 3.3 prophetic; Strand Life Sciences, San Francisco, CA, USA) was used for conducting microarray gene expression analysis. Data were imported as original.cel files from GCOS into the AVADIS software. Global normalisation was carried out using the Robust Multichip Averaging (RMA) algorithm (Irizarry et al. 2003). The dataset was filtered initially by removing all 100% absent calls (i.e. no signal) across all arrays, leaving 7916 genes that showed expression changes for further analysis. To confirm data normalisation, the dataset was viewed statistically by different methods such as heat maps, scatter plots and box plots.
Differential expression analysis was conducted individually for each time point, namely 4, 7 and 10 DAI for both inoculated and control treatments. Unpaired one-tailed statistical t-tests were used to determine the statistical significance (P ≤ 0.05) of gene expression. A fold cutoff value of 1.5 was used for filtering out significant genes. Two gene lists were generated for each time point; one for upregulated and one for downregulated genes. The probe set of up- and downregulated genes at each time point (4, 7 and 10 DAI) were saved as.txt files, and batch searches were conducted for gene annotation. Gene annotation was carried out by use of the NetAffx Analysis Centre (Affymetrix) (www.affymetrix.com/analysis) for functionality determinations. The results of these queries were exported as tab separated files (.tsv) and saved as Microsoft Excel files.
Metabolic pathway analysis
The metabolic pathways controlled by specific genes were identified using AraCyc (The Pathway Tools version 10.0 and AraCyc version 2.6, available from The Arabidopsis Information Resource Website (http://www.arabidopsis.org/tools/aracyc)). The AraCyc database for A. thaliana Col-0 was used for running the expression viewer tool at each time point. The log values from the tab-delimited files were used by AraCyc for displaying the reactions in a colour relative to the expression levels.
To obtain a more general overview of gene expression changes, analysis using MAPMAN (Thimm et al. 2004) was conducted on RMA normalised data, but without the cutoffs described above. Ratios of control to infected roots were calculated for each time point for all values and the values were converted to log2 in Microsoft Excel files.
Plant growth and inoculation of A. thaliana mutants
A. thaliana ecotype Col-0 and mutants jar1 (jasmonate resistant; allelic to jin4) (Staswick et al. 1992) (CS8072) and coi1 (coronatine insensitive) (Feys et al. 1994) (Salk_035548) seeds were sterilised in 50% (v : v) ethanol, 5% (v : v) hydrogen peroxide and 45% (v : v) sterile distilled water for 5 min with gentle agitation. The seeds were then rinsed three times with sterile distilled water to remove residual ethanol and hydrogen peroxide. Seeds were sown in Petri plates on Murashige and Skoog basal medium (Sigma-Aldrich, St Louis, MO, USA; Murashige and Skoog 1962) containing 1% sucrose, 0.8 g L–1 agar (Sigma) and pH adjusted to 5.8. Seeds were stratified at 4°C for 72 h in the dark and then incubated at 21°C with a 12-h photoperiod (100 µmol m–2 s–1). Seven-day-old seedlings were transferred from plates into trays containing soil mix. The soil mix consisted of a mixture of 30% (v : v) peat moss, 30% (v : v) propagating sand and 40% (v : v) vermiculite (size 2). Seventeen-day-old seedlings were inoculated as described above. Control plants were grown in separate trays and mock-inoculated. Screening of A. thaliana mutant lines required up to 25 to 30 plants per mutant. Disease was assessed at 31 DAI as described above.
Quantitative real-time reverse transcriptase PCR analysis
The same RNA samples used previously for the microarray experiment and stored at −80°C were used in PCR experiments as suggested by Rajeevan et al. (2001). RNA samples from two replicates from the control and treatment plants at 4 DAI were treated with DNase I (DNA-free™ Kit, Ambion), and total RNA (between 50 ng and 2 µg) was then reverse transcribed into cDNA (Omniscript Reverse Transcriptase Kit, Qiagen, Düesseldorf, Germany). These samples were then used for setting up PCR reactions for determining transcript levels using quantitative reverse transcriptase PCR (RT–qPCR) analysis. The PCR reaction was set up in a 25-µL volume containing 12.5 µL of SybrGreen (Qiagen), 1 µL each of forward and reverse primers (30 ng each), RNA-free water (Ambion) and 1 µL of the template cDNA. The cDNA samples were diluted to 1 : 10 and used in duplicates to set up PCR reactions. Cycling conditions were 95°C for 15 min of initial denaturation, followed by 45 cycles of amplification in a three-step procedure; 40 s at 95°C, 30 s at 55°C and 30 s at 72°C, and followed by a three-step cycle of product melting (60–92°C with a 30-s hold on the first and next steps). Gene specific primer sequences for all five genes, including the A. thaliana Actin 2 reference gene (At3g18789) was designed using the Primer 3 software tool version 0.4.0 (Rozen and Skaletsky 2000) and are listed in Table S1 (available as an Accessory Publication to this paper). The relative quantification of a target gene in comparison with a reference gene was performed according to the method described by Pfaffl (2001) to account for differences in efficiency between the reference gene and the gene of interest.
Results
Disease progression in the susceptible A. thaliana ecotype Col-0
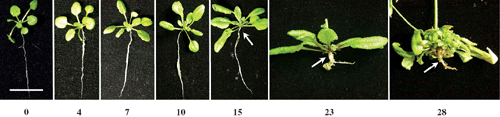
Following inoculation with P. brassicae, representative plants from each of the replicated experiments developed the characteristic symptoms of disease by 28 days. Plants were assessed for disease symptoms macroscopically at 4, 7, 10, 15, 23 and 28 DAI (Fig. 1). Root and shoot growth was normal at 4, 7 and 10 DAI with no apparent sign of the disease. Swelling in the hypocotyl was observed at 15 DAI, which developed into whitish root galls by 28 days. Lateral roots were completely destroyed and the whole plant was stressed, with a proliferation of multiple rosette leaves at 28 DAI. The timing and development of root galls were similar to that described previously (Agarwal et al. 2009).
|
Functional classification of differentially expressed genes at 4, 7 and 10 DAI
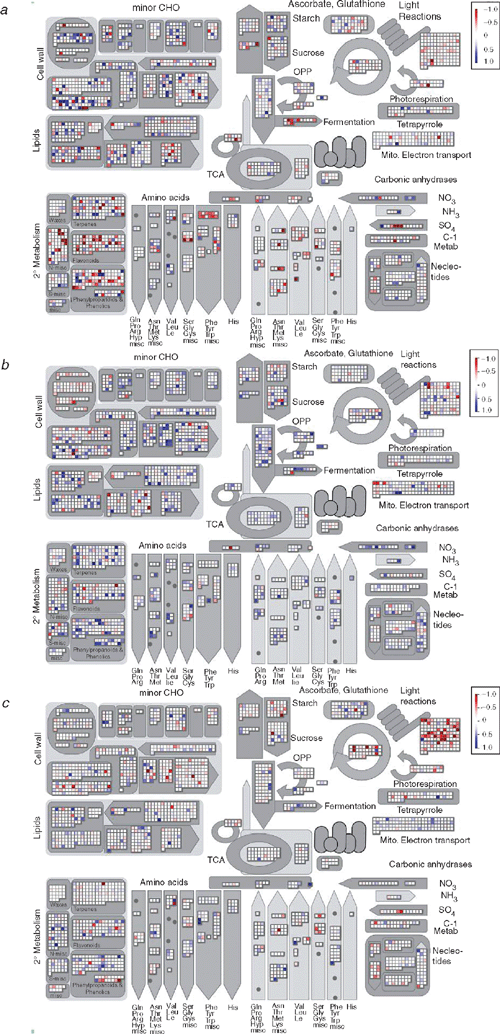
According to MAPMAN analysis, which was conducted with no fold change cutoff, the greatest change in expression of genes was apparent at 4 DAI (Fig. 2). When the data were analysed using a threshold of greater than a 1.5-fold increase or decrease above controls at a 95% confidence level, 147 genes were differentially expressed at 4 DAI as compared with 27 genes and 37 genes at 7 and 10 DAI, respectively (Tables S2–S7). At 4 DAI, 58 genes were upregulated compared with 22 and 31 genes 7 and 10 DAI, respectively. Similarly, 89 genes were found to be downregulated at 4 DAI, compared with 5 and 6 genes at 7 and 10 DAI, respectively. At each time point, the differentially expressed genes were classified into several functional categories including those that are stimulated by biotic and abiotic agents (response to stimuli); those involved in biosynthesis, metabolism, transport, signal transduction and transcription, cell wall modification, photosynthesis, cell death and cell adhesion; and also those of unknown function (Table 1). Different stages in disease progression correlated with different combinations of genes being induced or repressed.
|
Thirty-one out of 58 genes induced at 4 DAI (Table S2) were classified according to their known biological function. Twenty-one percent (12) of the genes induced were involved in signalling pathways. The genes that were upregulated at this time point included those that encoded a WRKY transcription factor and at least four kinases: calcium-dependent protein kinase (CDPK), a mitogen activated protein kinase kinase (MAPKK), serine–threonine protein kinase (PR5K) and leucine-rich repeat (LRR) protein kinase. Nine percent (5) of the genes were involved in the metabolism of various compounds including phenylpropanoids, lipids, carbohydrates and aromatics. Seven percent (4) of the genes induced were in response to stimuli such as pathogens (the disease resistance protein toll interleukin receptor – nucleotide binding site – LRR (TIR-NBS-LRR)), oxidative stress (peroxidase) and a hormone, GA. Another 7% (4) of the genes were involved in the transport of ions. Seven percent (4) of the genes involved in cell wall modification and cell growth, such as pectinesterase and expansin, were also induced at this time point. A lipoxygenase gene (LOX4) involved in the biosynthesis of jasmonic acid was upregulated by 3.8-fold. A single cell adhesion-related gene that encoded a fasciclin domain-containing protein was also induced. Forty-seven percent (27) of the induced genes were proteins with unknown function.
A higher number of genes were repressed rather than induced at 4 DAI in this host–pathogen interaction. Fifty-nine out of the 89 genes that were downregulated were classified into various functional groups (Table S3). Twenty-two percent (20) of the repressed genes were involved in cellular transport, including genes that encoded a MATE efflux protein, an oxidoreductase, an ABC transporter protein, a cytochrome P-450 and a glycine-rich protein. A further 16% (14) of the repressed genes were involved in biosynthesis pathways such as those for lignin, ethylene, aromatic amino acids, flavonoids and salicylic acid. A higher number of biosynthesis pathway genes (14) were downregulated, compared with only one upregulated biosynthesis pathway-related gene. Fifteen percent (13) of the downregulated genes were related to stimulus response. Genes involved in the oxidative burst pathway such as those that encode peroxidase, NADP-dependent oxidoreductase, heat-shock protein and glutathione S-transferase were all downregulated. An antimicrobial gene (encoding for chitinase) and a defence-related gene were also repressed. Seven percent (6) of the downregulated genes were involved in the metabolism of various compounds such as cytokinin. Only three of the genes involved in different signalling pathways were downregulated, compared with 12 genes that were upregulated. A zinc-finger family protein and one gene involved in cell growth, an expansin, were downregulated. The majority of the cell growth genes were upregulated. The fasciclin-like arabinogalactan protein, an adhesive molecule associated with cell walls, was downregulated. The chlorophyll a/b binding protein involved in the photosynthetic pathway was also downregulated. Thirty-four percent (30) of the suppressed genes were proteins with unknown function.
In comparison with the Day 4 time point, there were less number of genes that changed expression at 7 DAI, representing fewer functional categories. Out of the 22 genes induced (Table S4) and the 5 genes repressed (Table S5), only 15 and 4 genes were annotated, respectively. No enzyme involved in any of the biosynthesis pathways described above was induced or suppressed at this time point. Five of the genes induced were involved in the metabolic pathway of various compounds, four were involved in transport, two in response to stimuli, two in cell wall modification, two in signalling pathways and seven with unknown function. Only two genes of the genes involved in response to stimuli were repressed, one in signal transduction and transcription, one in cell death and one gene with unknown function.
Out of the 31 genes induced (Table S6) and the 6 genes repressed at 10 DAI (Table S7), only 15 and 5 genes, respectively, had known functions. Seven of the upregulated genes were involved in the signal transduction and transcription pathway. A zinc-finger protein and LRR protein kinase in this functional group were upregulated. Four of the genes were induced in response to stimuli, including the ABA-responsive protein, two genes were involved in metabolic pathways such as synthesis of cytokinin and another two genes were involved in biosynthesis pathways. For instance, 1-aminocyclopropane-1-carboxylate (ACC) oxidase gene, which is involved in the biosynthesis of ethylene, was induced. Fifty-two percent (16) of the genes could not be grouped under any known functional category. Two of the repressed genes that were downregulated were involved in the photosynthesis pathway, one gene was involved in the carbohydrate metabolism, one gene in response to stimuli, another one gene in the signal transduction and transcription pathway, and one had no function assigned to it.
Detailed analysis of gene regulation in metabolic pathways
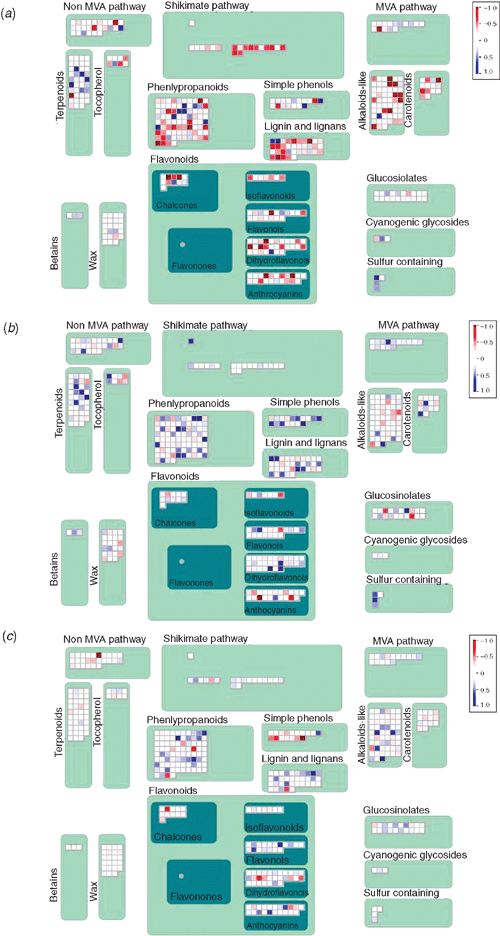
Following analysis of the gene expression patterns, it was evident that a large proportion of the differentially regulated genes were involved in metabolic pathways. In general, when comparing the MAPMAN plots, it becomes obvious that at 4 DAI, more genes were downregulated, both from general metabolism (Fig. 2) as well as from secondary metabolism (Fig. 3), whereas a greater proportion of genes were upregulated at 7 DAI. At the latest time point (10 DAI), there were fewer genes that showed expression changes, with the exception of genes related to photosynthesis, which were downregulated. This is reflected in the fact that the number of genes regulated in the metabolic pathways decreased over the time period. At 4 DAI, 23 up- and downregulated genes could be mapped on to the A. thaliana AraCyc metabolic pathway map, and only eight and four genes at 7 DAI and at 10 DAI, respectively (data not shown). These genes were involved in the biosynthesis of amino acids, co-factors, plant secondary metabolites, aromatic compounds, nucleosides and nucleotides, plant hormones, cell structures, fatty acids and lipids, photosynthesis; and in degradation, utilisation and assimilation.
|
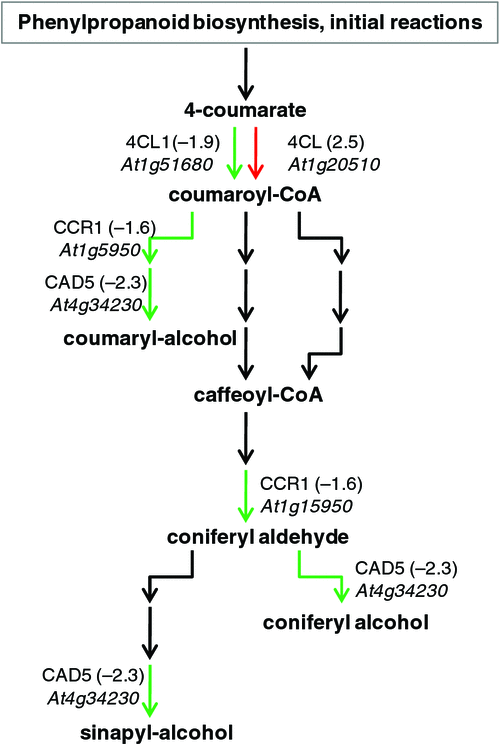
Phenylalanine ammonia-lyase catalyses the first step in the phenylproponoid biosynthesis pathway. A PAL4 gene (At3g10340) involved in initiation of one of the two possible salicylic acid biosynthesis pathways through the conversion of trans-cinnamate to (E)-cinnamoyl-coenzyme A was downregulated by 1.7-fold. The preceeding steps of the shikimate pathway (another salicylic acid biosynthesis pathway) were also downregulated at 4 DAI according to MAPMAN (Fig. S1). Furthermore, lignin biosynthesis was downregulated, as shown by the MAPMAN plots at 4 DAI, whereas the pathway was slightly upregulated in most genes at 7 and 10 DAI. According to AraCyc, at 4 DAI, genes involved in the synthesis of lignin; 4-coumarate-CoA ligase (4CL1, At1g51680), cinnamyl coenzyme A reductase (CCR1, At1g15950) and cinnamyl alcohol dehydrogenase (CAD5, At4g34230) were downregulated by 1.9-, 1.6- and 2.3-fold, respectively (Fig. 4).
|
Genes encoding the enzyme UDP-glucosyltransferase (At5g05860 and At2g30140), which were involved in the glycosylation of three cytokinins (cytokinin-0-glucoside, cytokinin 7-N-glucoside and cytokinin 9-N-glusoside) were both downregulated by 2.2-fold. In particular, all the reaction steps involved in the synthesis of cytokinin-7-N-glucoside starting from trans-zeatin to trans-zeatin-O-glucoside-7-N-glucoside were downregulated. The gene which encodes the enzyme 1-aminocyclopropane-1-carboxylate synthase (ACS2, At1g01480) that converts S-adenosyl-L-methionine to 1-aminocyclopropane-1-carboxylate in the ethylene biosynthesis pathway was downregulated by 1.7-fold. In the jasmonic acid biosynthesis pathway, only one gene encoding lipoxygenase (LOX4, At1g72520) was upregulated by 3.8-fold. The lipoxygenase enzyme controls the conversion of linolenate to 13(S)-hydroperoxy linolenate.
At 7 DAI, only eight upregulated genes could be mapped on to the AraCyc metabolic pathway. These genes were involved in the biosynthesis of amino acids, plant hormones, cell structures, sugars and polysaccharides, glycolysis and degradation/utilisation/assimilation.
At 10 DAI only four up- and downregulated genes could be mapped on to the AraCyc metabolic pathway. These genes were involved in the biosynthesis of plant hormones, fatty acids and lipids, photosynthesis; and in degradation, utilisation and assimilation. Interestingly, another gene encoding the enzyme UDP-glucosyltransferase (At3g53150) was upregulated by 1.9-fold at this time point.
Validation of microarray results – quantitative real-time RT–qPCR
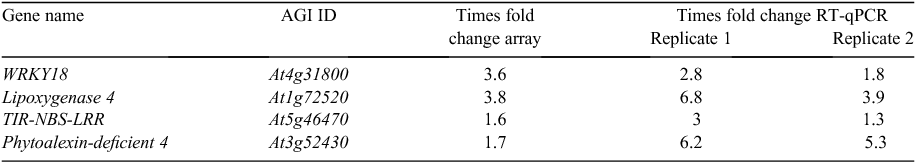
Microarray results were confirmed by validating the upregulation of four genes at 4 DAI (Table 2). The LOX4 gene, the WRKY transcription factor gene and the TIR-NBS-LRR disease resistance gene were all upregulated in the two replicates of RT–qPCR, postitively confirming the microarray data. For example, the LOX4 gene, which was highly upregulated by 3.8-fold according to the microarray results, showed expression level increases of 6.8- and 3.9-fold in the two RT–qPCR replicates.
Analysis of P. brassicae infection in A. thaliana mutants with impaired jasmonic acid signalling pathways and in Col-0 treated with salicylic acid
Two jasmonic acid signalling mutants, jar1 and coi1, were tested for resistance to clubroot. They were found to be susceptible to P. brassicae infection. The DI for both the mutants and Col-0 was equal to 100. There was no difference between the size of root galls and the shoots compared with the susceptible ecotype Col-0 (Fig. 5). All the inoculated plants developed root galls within 31 DAI.

|
As several genes related to salicylic acid biosynthesis were shown by microarray data to be downregulated at 4 DAI, the effect of treating plants with additional salicylic acid before inoculation was examined. Indeed, clubroot disease was strongly suppressed in A. thaliana plants infected with P. brassicae and treated with salicylic acid. The DI for the plants inoculated with P. brassicae and salicylic acid was 20 out of a possible 100. Plants inoculated with P. brassicae and and treated wtih salicylic acid were much healthier compared with non-inoculated but untreated plants, which had high levels of clubroot infection (Fig. 6) with a DI of 81.5. Fifty days after inoculation, only 50% of the treated plants inoculated with P. brassicae displayed visual symptoms of infection: 20% of the total number of plants had mild clubroot and 30% exhibited severe clubroot symptoms. Salicylic acid treated plants were, however, delayed in flowering, showing a reduction in the number of inflorescence stalks to two or three per plant compared with untreated control plants which produced four or five stalks per plant. In comparison, all the untreated plants inoculated with P. brassicae were severely infected, making them stunted in growth with multiple rosette leaves.

|
Discussion
This study describes the host gene expression changes that occur during the interaction between the susceptible A. thaliana Col-0 and P. brassicae at key developmental stages of the disease up to 10 DAI. Previous work has shown that the 4-day time point was critical for pathogen attachment and penetration in the host and for the onset of the disease (Agarwal et al. 2009). Quantitative real-time PCR (qPCR) analysis has also determined that the pathogen can first be detected in roots at 4 DAI in this system (Agarwal 2009). As a result, 4 DAI was selected as the first time point for gene expression analysis. Seven- and 10-day time points were also selected because they represent clearly different pathogen growth stages of the primary phase of the P. brassicae lifecycle within the host according to microscopic studies (Agarwal et al. 2009).
A relatively small subset of genes was differentially expressed in this study when using a biologically significant change cutoff of 1.5-fold. These findings are in agreement with the earlier work reported by Cao et al. (2008) where only a small subset of proteins with a low fold change were differentially identified at the early stages of host–pathogen interaction in susceptible canola. These results suggest that during the primary phase of the P. brassicae lifecycle, fewer morphological and physiological changes occur in the host compared with the secondary stage. Siemens et al. (2006) reported high numbers of differentially expressed genes at a greater fold change during the secondary stages of the disease, by which time the host roots exhibited systemic abnormalities. Also, since this study was conducted in a compatible host, it was expected that the changes in the plant would be subtle, as opposed to those in a resistant host, where the number of genes expressed are larger with higher fold changes. This phenomenon has been shown in other host–pathogen interactions (Bhattarai et al. 2008, for example) and similarly, a small subset of genes with low fold change has also been reported for the compatible interaction between Phytophthora infestans (Mont.) de Bary and Solanum tuberosum L. (Restrepo et al. 2005). In addition, it should be noted that root hair infection was monitored and that root hairs constitute only a small proportion of the white root tissue, maybe thus diluting the effect of the pathogen on differential gene expression.
Some of the genes known to be induced during defence responses in a resistant interaction are also induced during susceptible interactions, although with lower kinetics, as reported by Wise et al. (2007). It is also worth noting that the expression of defence-related genes is often temporally controlled very quickly during pathogen challenge and even a few hours can produce significant changes (Trusov et al. 2006; Rookes et al. 2008). Therefore, while the time points of 4, 7 and 10 DAI provide a good sample of gene expression in the interaction, it is possible that certain peaks or troughs in gene expression may be undetected. This is particularly relevant to pathways that may not exhibit uniform expression patterns.
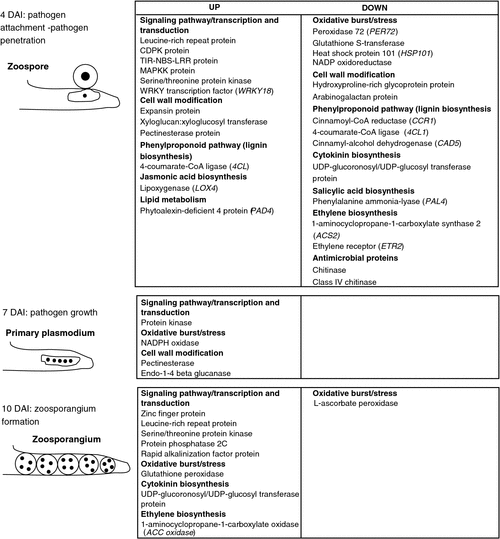
In a resistant interaction, recognition of a pathogen triggers several cellular events in plants and induces the early defence responses. These responses include activation of mitogen-activated protein kinases and NADPH oxidases, as well as the oxidative burst, which all occur within minutes of infection (Torres and Dangl 2005; Zhao et al. 2005; Jones and Dangl 2006). Transcription factors and early defence genes are also activated within the first few hours of the interaction. Generally, cascades of signalling molecules are triggered, including salicylic acid, jasmonic acid and ethylene (Schenk et al. 2000; Wiermer et al. 2005). Various defence-related proteins such as pathogenesis-related proteins with antimicrobial activity, glutathione S-transferases, peroxidases and proteinase inhibitors are activated, and the host is thus able to resist pathogen attack and is therefore resistant (Thatcher et al. 2005). Several of these factors were also found to be differentially regulated in the present study at 4 DAI when the first contact between the pathogen and host occurs. From these events, it can be concluded that some plant defence responses are transiently increased. Generally, biotrophs require a living host for nutrient acquisition and to complete their lifecycle. Unlike other biotrophs, however, P. brassicae does not form any apparent specialised structure, such as a haustorium, for this purpose – rather, after zoospore encystment, the protoplast from the encysted spore is injected into the host cytoplasm and plasmodia form within root hairs. Thus, the plasmodium is in continuous contact with the host cytoplasm, so it is likely that smaller changes in host metabolism are sufficient for pathogen nutrition. In addition, genes involved in cell wall metabolism and modification were also altered (mostly downregulated) in transcription at 4 DAI. At this time point, the highest number of differentially regulated genes was found, which reflects the importance of Day 4 as the time when infection is initiated (Fig. 7). At 7 DAI, metabolism-related genes, especially sugars and polysaccharides, were increased in expression, indicating that the pathogen is inside the root hair and is hijacking nutrients from the host. Finally, at 10 DAI, the lowest number of genes that had changed expression was found, which coincides with the stage of the lifecycle where the zoospores produced within zoosporangia leave the host cells.
|
In this study, differentially regulated genes were determined and a classification of genes according to functional group was conducted. An exploration of the significance of these genes and their potential role in the interaction is described below in more detail.
Signalling pathway-related genes
At all time points, there was a general trend of inducing the genes involved in signal transduction and transcription. The largest number of genes (12) involved in signalling pathways were upregulated at 4 DAI, indicating that there was some degree of recognition of the pathogen by the host. A member of the LRR family of R proteins (At5g12940) and a gene encoding a C2 domain-containing protein (At4g34150), both known to be involved in signal transduction and in pathogen recognition (Baker et al. 1997; Feys and Parker 2000; Glazebrook 2001; Jones and Dangl 2006), were induced at 4 DAI, confirming pathogen recognition by the host. Genes encoding a TIR-NBS-LRR disease resistance protein that mediates resistance signalling (Feys and Parker 2000; Glazebrook 2001; Wiermer et al. 2005) (At5g46470), a CDPK (At3g57530), a MAPKK (At5g56580) and a PR5K (At4g31800) were all induced at 4 DAI and not at later time points. Some of these signalling genes are known to be induced due to an early response in resistant host–pathogen interactions, during wounding and also during systemic acquired resistance (SAR) induction. The WRKY proteins are a superfamily of transcription factors (TF) reported to be switched on early after pathogen infection, in response to wounding, by signalling substances such as salicylic acid and other stresses (Dellagi et al. 2000; Eulgem et al. 2000; Cheong et al. 2002; Zheng et al. 2006; Eulgem and Somssich 2007). They are known to be induced by pathogen infection or penetration in both susceptible and resistant hosts (Wise et al. 2007). In this study, WRKY18 (At4g31800) was strongly upregulated at 4 DAI only and not at later time points, indicating pathogen recognition in this susceptible interaction. WRKY18 has been shown to be involved in defence against the necrotrophic pathogens Botrytis cinerea Pers. and Fusarium graminearum Schwabe, while conversely, A. thaliana WRKY18 mutants display enhanced resistance to the hemibiotroph Pseudomonas syringae pv. tomato (Xu et al. 2006; Chen et al. 2010; Makandar et al. 2010). The expression of this TF gene in the biotrophic interaction between A. thaliana and P. brassicae may therefore be facilitating susceptibility. Future testing of the response of the WRKY18 mutant in this interaction would be of great interest. Only two signal transduction-related genes were upregulated at 7 DAI, but at 10 DAI, this had increased to seven. This may be related to the lifecycle stage of P. brassicae, as the pathogen is only developing within the host root at 7 DAI, but at 10 DAI, the increase may be related to the mechanism of secondary zoospore release from the host root. This involves disruption of the host’s root hair plasma membrane and cell wall, causing considerable physiological changes to host cells, and also provides the potential for exposure of the pathogen to previously unaffected cells.
Genes involved in oxidative burst and stress
Recognition of a pathogen by the host plant is often followed by a burst of reactive oxygen intermediates (ROIs) (Baker et al. 1997; Schulze-Lefert 2004; Torres and Dangl 2005). In this study, most of the differentially regulated genes related to the oxidative burst such as peroxidase, glutathione S-transferase, heat shock proteins and NADP oxidoreductase genes were repressed at 4 DAI. Peroxidases and ROIs function not only as antimicrobial compounds but also as signalling molecules in plant defence (Wan et al. 2002). The data presented here indicate that ROIs are suppressed during pathogen penetration in the early stages of disease progression in the compatible interaction of P. brassicae with A. thaliana to facilitate proliferation of this biotrophic pathogen. Suppression of oxidative burst molecules has been reported in other compatible plant–pathogen interactions, such as in bean (Phaseolus vulgaris L.) leaf tissue challenged by B. cinerea (Unger et al. 2005). Differential expression of the proteins involved in oxidative stress was also reported in the proteomic study conducted by Devos et al. (2006) at the 4-day time point in A. thaliana roots inoculated with P. brassicae, and in the study by Cao et al. (2008) on canola roots.
Genes important for cell wall modification
Several genes involved in cell wall biosynthesis and modifications have been shown to be regulated by wounding and pathogen attack (Maleck et al. 2000; Schenk et al. 2000; Cheong et al. 2002). In this study, at 4 DAI (the approximate time point of pathogen penetration), several genes encoding enzymes such as expansin, pectin, xyloglucan endotransglycosylases, glycosyl transferases and pectinesterase, which are involved in cell wall modification, synthesis or both were upregulated. Similarly, in a study conducted by Devos et al. (2005) in Chinese cabbage (Brassica rapa var. pekinensis) plants susceptible to P. brassicae, there was an increase in xyloglucan activity in the epidermal root layer during the early infection stage. The expression of several genes whose protein products promote cell wall modification (e.g. expansins) may loosen the cell wall integrity, potentially making cells more vulnerable to pathogen ingress. On the other hand, the root hairs show swelling after infection during development of the plasmodia, so there could be additional stimuli for cell wall expansion.
Hydroxyproline-rich glycoprotein and arabinogalactan protein, which are important components of the plant cell wall (Estevez et al. 2006), were downregulated at the early time point. These proteins play a role in cell wall–plasma membrane cytoskeleton connections and are involved in plant detection of invading pathogens (Mellersh and Heath 2001; Hardham et al. 2007). It has been reported that a plant’s response to a pathogen is not accompanied by an increase in the level of hydroxyproline-rich glycoprotein in susceptible plants (Halverson and Stacey 1986), which supports the findings presented here.
An important plant defence response to pathogen attack and wounding is the activation of the phenylpropanoid pathway that produces many secondary metabolites (Cheong et al. 2002). Cell wall lignification, callose deposition and cell wall thickening are early defence responses, and reduce the spread of the pathogen in the host (Baker et al. 1997; Heath 1997; Mellersh and Heath 2001; Schulze-Lefert 2004; Thatcher et al. 2005). In this study, several genes encoding enzymes in the lignin biosynthesis pathway were suppressed at 4 DAI (Fig. 7). Strong suppression of the protein caffeoyl-CoA O-methyltransferase (CCoAOMT) in the lignin biosynthesis pathway was also reported by Cao et al. (2008) in the proteomic study of P. brassicae–B. napus interaction at 48 h after inoculation. Suppression of lignin biosynthesis pathway genes by ABA treatment has previously been shown to increase susceptibility in A. thaliana leaves infected with P. syringae (Mohr and Cahill 2007). Thickening and lignification of A. thaliana cell walls has been described in a histological study by Kobelt et al. (2000) in incompatible but not compatible interactions with P. brassicae, which supports the expression data presented here. Genes encoding the synthesis of β-1,3 glucan (callose) were also suppressed, further facilitating pathogen penetration in the P. brassicae compatible interaction. There is good evidence from other compatible host–pathogen interactions that host cell wall penetration is often accompanied by microbial suppression of host defences or cell death (Panstruga 2003; O’Connell and Panstruga 2006).
Genes involved in hormone synthesis, signalling and defence
The role of hormones such as jasmonic acid, ethylene, cytokinins and auxins during clubroot development has recently been reviewed by Ludwig-Müller and Schuller (2008). Several studies indicate that there is an increase in cytokinins at late time points during root gall formation (Devos et al. 2006; Siemens et al. 2006; Ludwig-Müller and Schuller 2008). The current microarray results confirm the involvement of cytokinins at the early infection stage. At 4 DAI, the synthesis of all three cytokinin glucosides (cytokinin-0-glucoside, cytokinin 7-N-glucoside and cytokinin 9-N-glucoside) in infected roots was suppressed but were induced 10 DAI. Since the cytokinin glucosides are considered inactive compounds, but can be hydrolysed to active cytokinins (Mok and Mok 2001), this might indicate that active cytokinin is needed at different time points during early interaction. Suppression of glycosylation would result in higher levels of cytokinins at 4 DAI, whereas an increase in glycosylation would diminish the levels of active cytokinin at 10 DAI. Devos et al. (2006) reported the onset of cytokinin response in the pericycle and the vascular tissue of A. thaliana 2 days after inoculation with P. brassicae using a cytokinin-inducible promoter::GUS construct. These findings support a possible role for cytokinins in early infection events. Transgenic plants with lower cytokinin levels were found to be more tolerant to clubroot (Siemens et al. 2006).
In this study, only one auxin-responsive protein was downregulated and one gibberellins-responsive protein upregulated at the 4-day time point. Only one ABA-responsive protein induced as a defence response was upregulated at 10 DAI and downregulated at 7 dAI. The data might indicate that these plant hormones, which are thought to be important for later infection stages (Siemens et al. 2006; Ludwig-Müller and Schuller 2008), are not important for the early phase of development of P. brassicae in A. thaliana.
Salicylic acid is an important phytohormone and has been shown to play critical roles in plant defence signalling (Dong 1998; Loake and Grant 2007; Leon-Reyes et al. 2010). Salicylic acid, either produced endogenously or exogenously applied, is a potent inducer of SAR and can protect plants against pathogens (Wan et al. 2002; Thatcher et al. 2005). Suppression of the salicylic acid pathway in A. thaliana induces susceptibility to many biotrophic and hemibiotrophic pathogens such as P. syringae (Mohr and Cahill 2007), so the data provided here indicating downregulation of salicylic acid biosynthesis pathways in the susceptible A. thaliana–P. brassicae interaction are of considerable interest. Gene expression studies of various hosts in their interactions with biotrophs such as Perenospora parasitica (Pers.) Tuls., Erysiphe spp. and Pseudomonas syringae pv. syringae support the notion that activation of salicylic acid signalling is important for resistance (Schulze-Lefert and Panstruga 2003; Oliver and Ipcho 2004; Glazebrook 2005; Zhao et al. 2005).
Ethylene and jasmonic acid (are important phytohormones involved in plant growth and development, and are also produced in response to both biotic and abiotic stresses (Schenk et al. 2000; Wan et al. 2002). Generally, ethylene and jasmonic acid responses are known to be important in defence against necrotrophic pathogens (Hammond-Kosack and Parker 2003; Oliver and Ipcho 2004; Glazebrook 2005; Zhao et al. 2005). A lipoxygenase gene (LOX4) that is important for jasmonic acid biosynthesis was strongly induced at 4 DAI, suggesting that jasmonic acid induction may be important for compatibility in P. brassicae interactions. Indeed, jasmonic acid may be acting as an antagonist of salicylic acid synthesis, as has been suggested by several researchers (Kunkel and Brooks 2002; Glazebrook et al. 2003). The jar1 mutant was more sensitive to clubroot infection, according to Siemens et al. (2002), and was also confirmed to be susceptible to the Australian population of P. brassicae. Induction of one lipoxygenase gene at 23 DAI has been shown by Siemens et al. (2006), indicating a potential role for jasmonic acid during gall development (Ludwig-Müller and Schuller 2008). In contrast, several genes involved in the ethylene pathway (1-aminocyclopropane-1-carboxylate synthase 2 and a putative ethylene receptor, ETR2) were downregulated at the early time point. As discussed in a recent review (Ludwig-Müller and Schuller 2008), according to the microarray analysis of Siemens et al. (2006), several ethylene responsive genes were differentially expressed, clearly indicating a role for ethylene in clubroot development. Interestingly, the ethylene-resistant mutants (etr1–1 and etr1–3) and ethylene-insensitive mutants (ein3–1 and ein4) tested by Siemens et al. (2002) did not show tolerance to clubroot. Resistance to P. brassicae infection was shown by Devos et al. (2006) in the alh1 mutant defective in cross-talk between ethylene and auxin. It is evident that there is considerable hormonal involvement in the A. thaliana–P. brassicae interaction at both the infection stage and during gall formation; however, there appears to be much complexity in the temporal control of various hormones during the various phases of disease. More detailed analysis of hormonal involvement in this interaction is still required.
Antimicrobial proteins, which include pathogenesis-related proteins such as chitinases and glucanases, are induced during stress, upon pathogen infection and following induction of SAR (Thatcher et al. 2005). In this study, two chitinase genes were suppressed at 4 DAI, which may reduce plant resistance to pathogen penetration.
Manipulation of defence hormone signalling to alter susceptibility to P. brassicae
The microarray analysis conducted in this study suggests that the biosynthesis of salicylic acid and ethylene may both be suppressed during early pathogen colonisation, but, in contrast, the biosynthesis of jasmonic acid is induced. As reported by Leon-Reyes et al. (2010), the interplay between salicylic acid, jasmonic acid and ethylene signalling is highly complex and is dependent on the sequence in which the hormones are produced. It is, however, well established in several plant–pathogen combinations that biotrophic interactions are controlled by salicylic acid-dependent defence pathways,whereas necrotrophic interactions are controlled by jasmonic acid- and ethylene-dependent defence pathways (Hammond-Kosack and Parker 2003; Oliver and Ipcho 2004; Glazebrook 2005). Therefore, it was of great interest to examine whether manipulation of these pathways could influence susceptibility to P. brassicae. Two jasmonic pathway mutants, jar1 and coi1, were found to exhibit similar levels of susceptibility as wild-type plants, indicating that this pathway is not particularly influential in the outcome of the interaction. In an extensive susceptibility screen conducted by Siemens et al. (2002) using P. brassicae isolate e3, the jar1 mutant was found to display enhanced susceptibility. Differences between pathogen isolates are a probable explanation for this discrepancy, but future testing to clarify this should be conducted.
In contrast, pretreatment of A. thaliana plants with salicylic acid was able to reduce the susceptibility to P. brassicae. This result, taken in context with the overall trend of either downregulation or a lack of induction of many genes implicated in the salicylic acid pathway in this interaction, suggests that amelioration of the pathway through either the addition of exogenous salicylic acid or pathway modifications through genetic means could provide effective strategies to increase resistance to P. brassicae in susceptible plant species. This is currently under further investigation.
Conclusion
The current research has provided further insights into the biology of P. brassicae during clubroot disease development, and provided information on the cellular and molecular changes that occur when the pathogen commences penetration of host cells and starts colonising root hairs. Importantly, this study has demonstrated that in the early stages of the interaction, repression of defence-related genes during invasion and colonisation by the pathogen appears to be necessary for the establishment of the pathogen within host roots. The expression levels of several genes known to be important for recognition and signal transduction in resistant interactions; and genes involved in general and secondary metabolism, cell wall modification, and lignin, ethylene, salicylic acid and cytokinin biosynthesis were downregulated. Furthermore, when salicylic acid was used as a potential SAR inducer, clubroot disease in A. thaliana was suppressed, indicating that manipulation of the salicylic acid signalling pathway may enable enhanced resistance to P. brassicae in the field.
Acknowledgements
This work has been funded by DPI Victoria and Horticulture Australia Limited (HAL) using the vegetable levy and matched funds from the Australian Government. The authors also thank DPI Victoria for providing access to facilities and Dr Caroline Donald for her critical review of the manuscript.
References
Agarwal A (2009) Interactions of Plasmodiophora brassicae with Arabidopsis thaliana. PhD thesis, Deakin University, Australia.Agarwal A, Kaul V, Faggian R, Cahill DM (2009) Development and use of a model system to monitor clubroot disease progression with an Australian field population of Plasmodiophora brassicae. Australasian Plant Pathology 38, 120–127.
| Development and use of a model system to monitor clubroot disease progression with an Australian field population of Plasmodiophora brassicae.Crossref | GoogleScholarGoogle Scholar |
Aist JR, Williams PH (1971) The cytology and kinetics of cabbage root hair penetration by Plasmodiophora brassicae Woron. Canadian Journal of Botany 49, 2023–2034.
| The cytology and kinetics of cabbage root hair penetration by Plasmodiophora brassicae Woron.Crossref | GoogleScholarGoogle Scholar |
Anderson JP, Thatcher LF, Singh KB (2005) Plant defense responses: conservation between models and crops. Functional Plant Biology 32, 21–34.
| Plant defense responses: conservation between models and crops.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXnt1Sltw%3D%3D&md5=4c1fd833b91d83bbd300064393a8f3d4CAS |
Baker B, Zambryski P, Staskawicz B, Dinesh-Kumar SP (1997) Signalling in plant–microbe interactions. Science 276, 726–733.
| Signalling in plant–microbe interactions.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK2sXivFOltLg%3D&md5=5cc9fabda636619e44cd04a26bdcfb30CAS | 9115193PubMed |
Bhattarai KK, Xie Q-G, Mantelin S, Bishnoi U, Girke T, Navarre DA, Kaloshian I (2008) Tomato susceptibility to root-knot nematodes requires an intact jasmonic acid signalling pathway. Molecular Plant-Microbe Interactions 21, 1205–1214.
Braselton JP (1995) Current status of the plasmodiophorids. Critical Reviews in Microbiology 21, 263–275.
| Current status of the plasmodiophorids.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DyaK287pt1Cjug%3D%3D&md5=01de44aebe8a4617264c0fd9a8821f71CAS | 8688155PubMed |
Cao T, Srivastava S, Rahman MH, Kav NNV, Hotte N, Deyholos MK, Strelkov SE (2008) Proteome-level changes in the roots of Brassica napus as a result of Plansmodiophora brassicae infection. Plant Science 174, 97–115.
Chen H, Lai Z, Shi J, Xiao Y, Chen Z, Xu X (2010) Roles of Arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress. BMC Plant Biology 10, 281
| Roles of Arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXmtFA%3D&md5=dc8c62551585308a9e7103b597e20c52CAS | 21167067PubMed |
Cheong YH, Chang H, Gupta R, Wang X, Zhu T, Luan S (2002) Transcriptional profiling reveals novel interactions between wounding, pathogen, abiotic stress, and hormonal responses in Arabidopsis. Plant Physiology 129, 661–677.
| Transcriptional profiling reveals novel interactions between wounding, pathogen, abiotic stress, and hormonal responses in Arabidopsis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38XkvV2jtrk%3D&md5=6214d91a5c6d1350f26cdb7aa7f4ed42CAS | 12068110PubMed |
Crute IR, Gray AR, Crisp P, Buczacki ST (1980) Variation in Plasmodiophora brassicae and resistance to clubroot disease in brassicas and allied crops – a critical review. Plant Breeding Abstracts 50, 91–104.
da Cunha L, Sreerekha MV, Mackey D (2007) Defense suppression by virulence effectors of bacterial phytopathogens. Current Opinion in Plant Biology 10, 349–357.
| Defense suppression by virulence effectors of bacterial phytopathogens.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXosFGrsLc%3D&md5=bab21a3a0ec29fff83db09f8504436afCAS | 17625953PubMed |
Dekhuijzen HM (1979) Electron microscopic studies on the root hairs and cortex of a susceptible and a resistant variety of Brassica campestris infected with Plasmodiophora brassicae. Netherlands Journal of Plant Pathology 85, 1–17.
| Electron microscopic studies on the root hairs and cortex of a susceptible and a resistant variety of Brassica campestris infected with Plasmodiophora brassicae.Crossref | GoogleScholarGoogle Scholar |
Dellagi A, Heilbronn J, Avrova AO, Montesano M, Palva ET, Stewart HE, Toth IK, Cooke DEL, Lyon GD, Birch PRJ (2000) A potato gene encoding a WRKY-like transcription factor is induced in interactions with Erwinia carotovora subsp. atroseptica and Phytophthora infestans and is coregulated with class 1 endochitinase expression. Molecular Plant—Microbe Interactions 13, 1092–1101.
| A potato gene encoding a WRKY-like transcription factor is induced in interactions with Erwinia carotovora subsp. atroseptica and Phytophthora infestans and is coregulated with class 1 endochitinase expression.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3cXntVarsrs%3D&md5=f86e4bcf7c26084a0f4a10c7a171ad91CAS | 11043470PubMed |
Devos S, Vissenberg K, Verbelen JP, Prinsen E (2005) Infection of Chinese cabbage by Plasmodiophora brassicae leads to a stimulation of plant growth: impacts on cell wall metabolism and hormone balance. New Phytologist 166, 241–250.
| Infection of Chinese cabbage by Plasmodiophora brassicae leads to a stimulation of plant growth: impacts on cell wall metabolism and hormone balance.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXjtlaku7g%3D&md5=585f306bbf52f6f3ad3267e255918093CAS | 15760367PubMed |
Devos S, Laukens K, Deckers P, Straeten DVD, Beeckman T, Inze D, Onckelen HV, Witters P, Prinsen E (2006) A hormone and proteome approach to picturing the initial metabolic events during Plasmodiophora brassicae infection on Arabidopsis. Molecular Plant—Microbe Interactions 19, 1431–1443.
| A hormone and proteome approach to picturing the initial metabolic events during Plasmodiophora brassicae infection on Arabidopsis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28Xht1yjs7rO&md5=257f22597b85239e10a4dd8ecf93e8abCAS | 17153927PubMed |
Dixon GR (2009) The occurrence and economic impact of Plasmodiophora brassicae and clubroot disease. Journal of Plant Growth Regulation 28, 194–202.
| The occurrence and economic impact of Plasmodiophora brassicae and clubroot disease.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXhtVSisbvL&md5=a0a3ba198e749fa41abe8bd4d6c8ad21CAS |
Donald C, Porter I (2009) Integrated control of clubroot. Journal of Plant Growth Regulation 28, 289–303.
| Integrated control of clubroot.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXhtVSisbjO&md5=239687368e1b58d48c8ad57a02b2c59bCAS |
Donald EC, Jaudzens G, Porter IJ (2008) Pathology of cortical invasion by Plasmodiophora brassicae in clubroot resistant and susceptible Brassica oleracea hosts. Plant Pathology 33, 585–589.
Dong X (1998) SA, JA, ethylene, and disease resistance in plants. Current Opinion in Plant Biology 1, 316–323.
| SA, JA, ethylene, and disease resistance in plants.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1cXls1Kktrc%3D&md5=36ed7661b43926241400e607a5c8d621CAS | 10066607PubMed |
Estevez JM, Kieliszewski MJ, Khitrov N, Somerville C (2006) Characterization of synthetic hydroxyproline-rich proteoglycans with arabinogalactan protein and extensin motifs in Arabidopsis. Plant Physiology 142, 458–470.
| Characterization of synthetic hydroxyproline-rich proteoglycans with arabinogalactan protein and extensin motifs in Arabidopsis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XhtFarsbnK&md5=ae290f6ac1a3b0bbf73a46d5826892eaCAS | 16935991PubMed |
Eulgem T, Somssich IE (2007) Networks of WRKY transcription factors in defense signalling. Current Opinion in Plant Biology 10, 366–371.
| Networks of WRKY transcription factors in defense signalling.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXosFGrsb8%3D&md5=cef975a0e5697504292b5465b698f2bfCAS | 17644023PubMed |
Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends in Plant Science 5, 199–206.
| The WRKY superfamily of plant transcription factors.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BD3c3kvFalsw%3D%3D&md5=70d6abb3483b858a339410281f9dc395CAS | 10785665PubMed |
Feys BJ, Parker JE (2000) Interplay of signalling pathways in plant disease resistance. Trends in Genetics 16, 449–455.
| Interplay of signalling pathways in plant disease resistance.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3cXotVSrsLc%3D&md5=9141807d465d580087ad163288baddc9CAS | 11050331PubMed |
Feys B, Benedetti CE, Penfold CN, Turner JG (1994) Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. The Plant Cell 6, 751–759.
Glazebrook J (2001) Genes controlling expression of defense responses in Arabidopsis – 2001 status. Current Opinion in Plant Biology 4, 301–308.
| Genes controlling expression of defense responses in Arabidopsis – 2001 status.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3MXksVykt7c%3D&md5=a4983be058620c7bbe5ee9b2b608625eCAS | 11418339PubMed |
Glazebrook J (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annual Review of Phytopathology 43, 205–227.
| Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXhtVOksrrN&md5=24b1f4ae6d066cd8397bbb74846acb69CAS | 16078883PubMed |
Glazebrook J, Chen W, Estes B, Chang HS, Nawrath C, Metraux JP, Zhu T, Katagiri F (2003) Topology of the network integrating salicylate and jasmonate signal transduction derived from global expression phenotyping. The Plant Journal 34, 217–228.
| Topology of the network integrating salicylate and jasmonate signal transduction derived from global expression phenotyping.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3sXjvFOgt7g%3D&md5=f986225cab9e5819060fb370468dc894CAS | 12694596PubMed |
Halverson LJ, Stacey G (1986) Signal exchange in plant–microbe interactions. Microbiological Reviews 50, 193–225.
Hammond-Kosack KE, Parker JE (2003) Deciphering plant–pathogen communication: fresh perspectives for molecular resistance breeding. Current Opinion in Biotechnology 14, 177–193.
| Deciphering plant–pathogen communication: fresh perspectives for molecular resistance breeding.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3sXjsVGlu78%3D&md5=6244034965e20b2ce08a50363a0882d1CAS | 12732319PubMed |
Hardham AR, Jones DA, Takemoto D (2007) Cytoskeleton and cell wall function in penetration resistance. Current Opinion in Plant Biology 10, 342–348.
| Cytoskeleton and cell wall function in penetration resistance.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXosFGrsLY%3D&md5=4c023cbed9cbaf6e0ccfa33ded4c4291CAS | 17627866PubMed |
Heath MC (1997) Signalling between pathogenic rust fungi and resistant or susceptible host plants. Annals of Botany 80, 713–720.
| Signalling between pathogenic rust fungi and resistant or susceptible host plants.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1cXlt1er&md5=56b3c6de49ab5194b7009762bd700615CAS |
Hirai M (2006) Genetic analysis of clubroot resistance in Brassica crops. Breeding Science 56, 223–229.
| Genetic analysis of clubroot resistance in Brassica crops.Crossref | GoogleScholarGoogle Scholar |
Ingram DS, Tommerup IC (1972) The life history of Plasmodiophora brassicae Woron. Proceedings of the Royal Society of London 180, 103–112.
| The life history of Plasmodiophora brassicae Woron.Crossref | GoogleScholarGoogle Scholar |
Irizarry RA, Hobbs B, Collin F, Bazer-Barclay YD, Antonrllis KJ, Scherf U, Speed TP (2003) Exploration, normalization and summaries of high density oligonucleotide array probe level data. Biostatistics 4, 249–264.
| Exploration, normalization and summaries of high density oligonucleotide array probe level data.Crossref | GoogleScholarGoogle Scholar | 12925520PubMed |
Jones JD, Dangl JL (2006) The plant immune system. Nature 444, 323–329.
| The plant immune system.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28Xht1SgtbzO&md5=289500416c19947efd96d3e4aedb7bf5CAS | 17108957PubMed |
Jones DR, Ingram DS, Dixon GR (1982) Characterisation of isolates derived from single resting spores of Plasmodiophora brassicae and studies of their interaction. Plant Pathology 31, 239–246.
| Characterisation of isolates derived from single resting spores of Plasmodiophora brassicae and studies of their interaction.Crossref | GoogleScholarGoogle Scholar |
Kobelt P, Siemens J, Sacristan MD (2000) Histological characterisation of the incompatible interaction between Arabidopsis thaliana and the obligate biotrophic pathogen Plasmodiophora brassicae. Mycological Research 104, 220–225.
| Histological characterisation of the incompatible interaction between Arabidopsis thaliana and the obligate biotrophic pathogen Plasmodiophora brassicae.Crossref | GoogleScholarGoogle Scholar |
Kunkel BN, Brooks DM (2002) Cross talk between signalling pathways in pathogen defense. Current Opinion in Plant Biology 5, 325–331.
| Cross talk between signalling pathways in pathogen defense.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38Xls1ejtLs%3D&md5=14244877663fc48590afa26bf6ad63f3CAS | 12179966PubMed |
Leon-Reyes A, Du Y, Koorneef A, Proletti S, Korbes AP, Memelink J, Pieterse Corne MJ, Ritsema T (2010) Ethylene signalling renders the jasmonate response of Arabidopsis insensitive to future suppression by salicylic acid. Molecular Plant—Microbe Interactions 23, 187–197.
| Ethylene signalling renders the jasmonate response of Arabidopsis insensitive to future suppression by salicylic acid.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXovVWktg%3D%3D&md5=a04b20ec69bee9944cd15bbc5d660beeCAS | 20064062PubMed |
Loake G, Grant M (2007) Salicylic acid in plant defense – the players and protagonists. Current Opinion in Plant Biology 10, 466–472.
| Salicylic acid in plant defense – the players and protagonists.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXhtFekurjM&md5=ab1e160be0d214a7ec681367d9be164dCAS | 17904410PubMed |
Ludwig-Müller J, Schuller A (2008) What can we learn from clubroots: alterations in host roots and hormone homeostasis caused by Plasmodiophora brassicae. European Journal of Plant Pathology 121, 291–302.
| What can we learn from clubroots: alterations in host roots and hormone homeostasis caused by Plasmodiophora brassicae.Crossref | GoogleScholarGoogle Scholar |
Makandar R, Nalam V, Chaturvedi R, Jeannotte R, Sparks AA, Shah J (2010) Involvement of salicylate and jasmonate signalling pathways in Arabidopsis interaction with Fusarium graminearum. Molecular Plant—Microbe Interactions 23, 861–870.
| Involvement of salicylate and jasmonate signalling pathways in Arabidopsis interaction with Fusarium graminearum.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXnvVart78%3D&md5=5b9b3133a7cfdfe00a878794db04cc7bCAS | 20521949PubMed |
Maleck K, Levine A, Eulgem T, Morgan A, Schmid J, Lawton KA, Dangl JL, Dietrich RA (2000) The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nature 26, 403–410.
Mellersh DG, Heath MC (2001) Plasma membrane–cell wall adhesion is required for expression of plant defense responses during fungal penetration. The Plant Cell 13, 413–424.
Mohr PG, Cahill DM (2007) Suppression of ABA of salicylic acid and lignin accumulation and the expression of multiple genes, in Arabidopsis infected with Pseudomonas syringae pv. Tomato. Functional & Integrative Genomics 7, 181–191.
| Suppression of ABA of salicylic acid and lignin accumulation and the expression of multiple genes, in Arabidopsis infected with Pseudomonas syringae pv. Tomato.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXls1SitLw%3D&md5=17280656328db8df35c0db7cc2617ac8CAS | 17149585PubMed |
Mok DWS, Mok MC (2001) Cytokinin metabolism and action. Annual Review of Plant Physiology and Plant Molecular Biology 52, 89–118.
| Cytokinin metabolism and action.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3MXkslWgsL8%3D&md5=41e6d9225c62d01294783894f27baae5CAS | 11337393PubMed |
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassay with tobacco tissue culture. Physiologia Plantarum 15, 473–497.
| A revised medium for rapid growth and bioassay with tobacco tissue culture.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaF3sXksFKm&md5=06c472aaff717f5157aa29da67ee5ad0CAS |
O’Connell RJ, Panstruga R (2006) Tete a tete inside a plant cell: establishing compatibility between plants and biotrophic fungi and oomycetes. New Phytologist 171, 699–718.
| Tete a tete inside a plant cell: establishing compatibility between plants and biotrophic fungi and oomycetes.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XhtVSku77F&md5=17649bfab9af6e31b1b71807f5e3829aCAS | 16918543PubMed |
Oliver RP, Ipcho SVS (2004) Arabidopsis pathology breathes new life into the necrotrophs-vs.-biotrophs classification of fungal pathogens. Molecular Plant Pathology 5, 347–352.
| Arabidopsis pathology breathes new life into the necrotrophs-vs.-biotrophs classification of fungal pathogens.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXmsl2ns7w%3D&md5=e0ca5da769490c60dbc69ac2f5d5c2c9CAS | 20565602PubMed |
Panstruga R (2003) Establishing compatibility between plants and obligate biotrophic pathogens. Current Opinion in Plant Biology 6, 320–326.
| Establishing compatibility between plants and obligate biotrophic pathogens.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3sXlsValt7c%3D&md5=12060abb1600a2dde9757c4aa3a931cdCAS | 12873525PubMed |
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Research 29, e45
| A new mathematical model for relative quantification in real-time RT–PCR.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BD38nis12jtw%3D%3D&md5=0957e876af3eab3c669ce09b7afcfd19CAS | 11328886PubMed |
Rajeevan MS, Vernon SD, Tayranang N, Unger ER (2001) Validation of array-based gene expression profiles by real-time (kinetic) RT-PCR. The Journal of Molecular Diagnostics 3, 26–31.
| Validation of array-based gene expression profiles by real-time (kinetic) RT-PCR.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3MXhsFahs7Y%3D&md5=4af53c03056c3c06d9674bdb6eb954e1CAS | 11227069PubMed |
Restrepo S, Myers KL, del Pozo O, Martin GB, Hart AL, Buell CR, Fry WE, Smart CD (2005) Gene profiling of a compatible interaction between Phytophthora infestans and Solanum tuberosum suggests a role for carbonic anhydrase. Molecular Plant—Microbe Interactions 18, 913–922.
| Gene profiling of a compatible interaction between Phytophthora infestans and Solanum tuberosum suggests a role for carbonic anhydrase.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXpsFGju7Y%3D&md5=79a2c0b12cf6e172cca1ced2f994732aCAS | 16167762PubMed |
Rookes JE, Wright ML, Cahill DM (2008) Elucidation of defense responses and signalling pathways induced in Arabidopsis thaliana following challenge with Phytophthora cinnamomi. Physiological and Molecular Plant Pathology 72, 151–161.
| Elucidation of defense responses and signalling pathways induced in Arabidopsis thaliana following challenge with Phytophthora cinnamomi.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXhsVKls7fM&md5=320806b507cda4234f5566157c115bc9CAS |
Rozen S, Skaletsky HJ (2000) Primer 3 on the WWW for general users and for biologist programmers. In: ‘Bioinformatics methods and protocols: methods in molecular biology’. (Eds S Krawetz, S Misener) pp. 365–386. (Humana Press: Totowa, NJ)
Schenk PM, Kazan K, Wilson I, Anderson JP, Richmond T, Somerville SC, Manners JM (2000) Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proceedings of the National Academy of Sciences of the United States of America 97, 11 655–11 660.
| Coordinated plant defense responses in Arabidopsis revealed by microarray analysis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3cXnsF2qtro%3D&md5=284bd9bb9001bcd89332bdd2da1dea63CAS |
Schulze-Lefert P (2004) Knocking on the heaven’s wall: pathogenesis of and resistance to biotrophic fungi at the cell wall. Current Opinion in Plant Biology 7, 377–383.
| Knocking on the heaven’s wall: pathogenesis of and resistance to biotrophic fungi at the cell wall.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXlsVWhtLw%3D&md5=f956f020b740c45a7714f3dc55c4e38fCAS | 15231259PubMed |
Schulze-Lefert P, Panstruga R (2003) Establishment of biotrophy by parasitic fungi and reprogramming of host cells for disease resistance. Annual Review of Phytopathology 41, 641–667.
| Establishment of biotrophy by parasitic fungi and reprogramming of host cells for disease resistance.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3sXptFWltr0%3D&md5=717b266d7466f985d8a285b7a5aa90eaCAS | 14527335PubMed |
Siemens J, Nagel M, Ludwig-Müller J, Sacristan MD (2002) The interaction of Plasmodiophora brassicae and Arabidopsis thaliana: parameters for disease quantification and screening of mutant lines. Journal of Phytopathology 150, 592–605.
| The interaction of Plasmodiophora brassicae and Arabidopsis thaliana: parameters for disease quantification and screening of mutant lines.Crossref | GoogleScholarGoogle Scholar |
Siemens J, Keller I, Sarx J, Kunz S, Schuller A, Nagel W, Schmulling T, Parniske M, Ludwig-Müller J (2006) Transcriptome analysis of Arabidopsis clubroots indicate a key role for cytokinins in disease development. Molecular Plant—Microbe Interactions 19, 480–494.
| Transcriptome analysis of Arabidopsis clubroots indicate a key role for cytokinins in disease development.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XktVClsL8%3D&md5=225c70c818f0958357bf7341219cf286CAS | 16673935PubMed |
Staswick PE, Su W, Howell SH (1992) Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proceedings of the National Academy of Sciences of the United States of America 89, 6837–6840.
Takahashi H, Ishikawa T, Kaido M, Takita K, Hayakawa T, Okazaki K, Itoh K, Mitsui T, Hori H (2006) Plasmodiophora brassicae-induced cell death and medium alkalization in clubroot-resistant cultured roots of Brassica rapa. Journal of Phytopathology 154, 156–162.
| Plasmodiophora brassicae-induced cell death and medium alkalization in clubroot-resistant cultured roots of Brassica rapa.Crossref | GoogleScholarGoogle Scholar |
Thatcher LF, Anderson JP, Singh KB (2005) Plant defence responses: what we have learnt from Arabidopsis? Functional Plant Biology 32, 1–19.
| Plant defence responses: what we have learnt from Arabidopsis?Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXpt1CksA%3D%3D&md5=30c5c9eff3345e12313f0958503ea986CAS |
Thimm O, Blaesing O, Gibon Y, Nagel A, Meyer S, Krueger P, Selbig J, Mueller LA, Rhee SY, Stitt M (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. The Plant Journal 37, 914–939.
| MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXjtFChu78%3D&md5=c2f785f57c6d48085d9cf7a5035146deCAS | 14996223PubMed |
Torres MA, Dangl JL (2005) Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Current Opinion in Plant Biology 8, 397–403.
| Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXlsFGgtrg%3D&md5=e1258106ee77df6e695bb9c181ebbe3bCAS | 15939662PubMed |
Toxopeus H, Dixon GR, Mattusch P (1986) Physiologic specialisation in Plasmodiophora brassicae: an analysis by international experimentation. Transactions of the British Mycological Society 87, 279–287.
| Physiologic specialisation in Plasmodiophora brassicae: an analysis by international experimentation.Crossref | GoogleScholarGoogle Scholar |
Trusov Y, Rookes JE, Chakravorty D, Armour D, Schenk PM, Botella JR (2006) Heterotrimeric G proteins facilitate Arabidopsis resistance to necrotrophic pathogens and are involved in jasmonate signaling. Plant Physiology 140, 210–220.
| Heterotrimeric G proteins facilitate Arabidopsis resistance to necrotrophic pathogens and are involved in jasmonate signaling.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XhtVCgsbY%3D&md5=0b3faed151b673944c503fb5a6daa01dCAS | 16339801PubMed |
Unger C, Kleta S, Jandl G, Tiedemann AV (2005) Suppression of the defense-related oxidative burst in bean leaf tissue and bean suspension cells by the necrotrophic pathogen Botrytis cinerea. Journal of Phytopathology 153, 15–26.
| Suppression of the defense-related oxidative burst in bean leaf tissue and bean suspension cells by the necrotrophic pathogen Botrytis cinerea.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXhvVCktbg%3D&md5=b88f7b09be14b80637e9ad05786b539cCAS |
Wan J, Dunning FM, Bent AF (2002) Probing plant–pathogen interactions and downstream defense signalling using DNA microarrays. Functional & Integrative Genomics 2, 259–273.
| Probing plant–pathogen interactions and downstream defense signalling using DNA microarrays.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38XovVWns78%3D&md5=cedcc0ee1b574553b3ad98815fb6fa19CAS | 12444419PubMed |
Wiermer M, Feys BJ, Parker JE (2005) Plant immunity: the EDS1 regulatory node. Current Opinion in Plant Biology 8, 383–389.
| Plant immunity: the EDS1 regulatory node.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXlsFGgtro%3D&md5=f3554e11686406e735bf182cc9fa4c7cCAS | 15939664PubMed |
Wise RP, Moscou MJ, Bogdanove AJ, Whitham SA (2007) Transcrpt profiling in host–pathogen interactions. Annual Review of Phytopathology 45, 329–369.
| Transcrpt profiling in host–pathogen interactions.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXhtVKls7vP&md5=e21dea8a8002a518a3cbdd35a3d89446CAS | 17480183PubMed |
Xu X, Chen C, Fan B, Chen Z (2006) Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. The Plant Cell 18, 1310–1326.
| Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XkslCmu7c%3D&md5=6bffffcb4dd43b29f33845df3adc367cCAS | 16603654PubMed |
Zhao J, Davis LC, Verpoorte R (2005) Elicitor signal transduction leading to production of plant secondary metabolites. Biotechnology Advances 23, 283–333.
| Elicitor signal transduction leading to production of plant secondary metabolites.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXjsVehu7c%3D&md5=42748ccafff0e90e4d513961d2845905CAS | 15848039PubMed |
Zheng Z, Qamar SA, Chen Z, Mengiste T (2006) Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. The Plant Journal 48, 592–605.
| Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28Xht1ylurfI&md5=32a7500a2f061fa5f0a3b670db33d6b8CAS | 17059405PubMed |